3VML
 
 | |
3VP9
 
 | | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, General transcriptional corepressor TUP1 | | Authors: | Matsumura, H, Kusaka, N, Nakamura, T, Tanaka, N, Sagegami, K, Uegaki, K, Inoue, T, Mukai, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications
J.Biol.Chem., 287, 2012
|
|
3AUA
 
 | | Crystal structure of the quaternary complex-2 of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[ethanoyl(hydroxy)amino]propylphosphonic acid, CALCIUM ION, ... | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
3VL7
 
 | |
1DR0
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD708 | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
1DR8
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD177 | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
6JTC
 
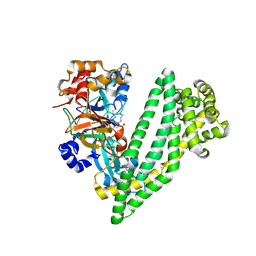 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with SH-5 from Porphyromonas gingivalis (Space) | | Descriptor: | 2-(2-azanylethylamino)-5-nitro-benzoic acid, Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
1DPZ
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD711 | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | Deposit date: | 1999-12-29 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
3VMK
 
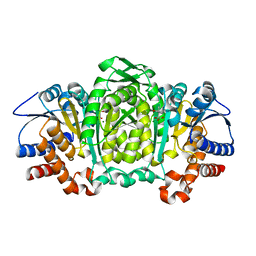 | | 3-isopropylmalate dehydrogenase from Shewanella benthica DB21 MT-2 | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, CHLORIDE ION, ... | | Authors: | Nagae, T, Watanabe, N. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural analysis of 3-isopropylmalate dehydrogenase from the obligate piezophile Shewanella benthica DB21MT-2 and the nonpiezophile Shewanella oneidensis MR-1
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3AU8
 
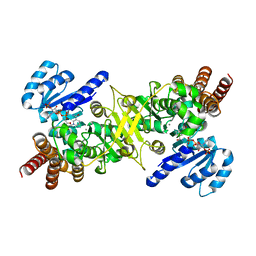 | | Crystal structure of the ternary complex of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
3AU9
 
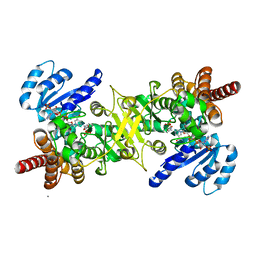 | | Crystal structure of the quaternary complex-1 of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CALCIUM ION, ... | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
3WVH
 
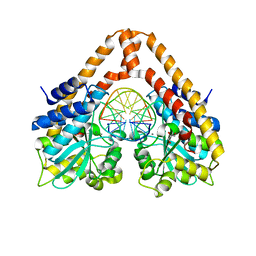 | | Time-Resolved Crystal Structure of HindIII with 25sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-05-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WVI
 
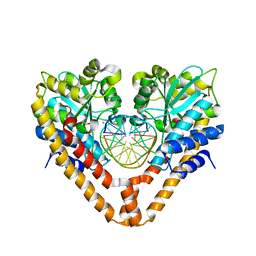 | | Time-Resolved Crystal Structure of HindIII with 40 sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-05-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2D2X
 
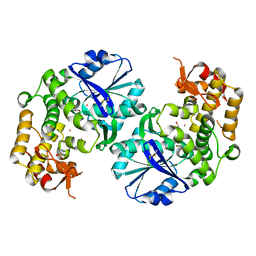 | | Crystal structure of 2-deoxy-scyllo-inosose synthase | | Descriptor: | 2-deoxy-scyllo-inosose synthase, COBALT (II) ION, GLYCEROL, ... | | Authors: | Nango, E, Kumasaka, T, Tanaka, N, Kakinuma, K, Eguchi, T. | | Deposit date: | 2005-09-20 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of 2-deoxy-scyllo-inosose synthase, a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics, in complex with a mechanism-based inhibitor and NAD+
Proteins, 70, 2008
|
|
3VMJ
 
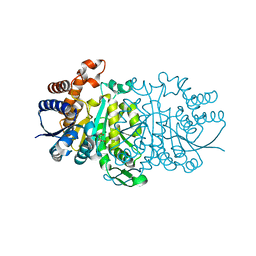 | | 3-isopropylmalate dehydrogenase from Shewanella oneidensis MR-1 | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, CHLORIDE ION, ... | | Authors: | Nagae, T, Watanabe, N. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural analysis of 3-isopropylmalate dehydrogenase from the obligate piezophile Shewanella benthica DB21MT-2 and the nonpiezophile Shewanella oneidensis MR-1
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3WVP
 
 | | Time-Resolved Crystal Structure of HindIII with 60sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*A)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), DNA (5'-D(P*AP*GP*CP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-06-02 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5YP2
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | Descriptor: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP3
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, ISOLEUCINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5Z98
 
 | | Crystal Structure of the Primate APOBEC3H Dimer mediated by RNA Duplex | | Descriptor: | Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like protein 3H, RNA (5'-R(*AP*UP*AP*CP*CP*CP*GP*GP*CP*A)-3'), RNA (5'-R(P*CP*UP*GP*CP*CP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Matsuoka, T, Nagae, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA.
Nucleic Acids Res., 46, 2018
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
3WOR
 
 | | Crystal structure of the DAP BII octapeptide complex | | Descriptor: | Angiotensin II, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOP
 
 | | Crystal structure of the DAP BII hexapeptide complex II | | Descriptor: | Angiotensin IV, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOQ
 
 | | Crystal structure of the DAP BII hexapeptide complex III | | Descriptor: | Angiotensin IV, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WON
 
 | | Crystal structure of the DAP BII dipeptide complex III | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOL
 
 | | Crystal structure of the DAP BII dipeptide complex I | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
