1WDY
 
 | | Crystal structure of ribonuclease | | Descriptor: | 2-5A-dependent ribonuclease, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Goto, Y, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-05-19 | | Release date: | 2004-10-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of 2',5'-linked oligoadenylates by human ribonuclease L
Embo J., 23, 2004
|
|
8WY1
 
 | | The structure of cyclization domain in cyclic beta-1,2-glucan synthase from Thermoanaerobacter italicus | | Descriptor: | Glycosyltransferase 36 | | Authors: | Tanaka, N, Saito, R, Kobayashi, K, Nakai, H, Kamo, S, Kuramochi, K, Taguchi, H, Nakajima, M, Masaike, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Functional and structural analysis of a cyclization domain in a cyclic beta-1,2-glucan synthase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
1WLT
 
 | | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue from Sulfolobus tokodaii | | Descriptor: | 176aa long hypothetical dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Watanabe, N, Sakai, N, Zilian, Z, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of dTDP-4-dehydrorhamnose 3,5-epimerase homologue
to be published
|
|
1FGJ
 
 | | X-RAY STRUCTURE OF HYDROXYLAMINE OXIDOREDUCTASE | | Descriptor: | HEME C, HYDROXYLAMINE OXIDOREDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tanaka, N, Igarashi, N, Moriyama, H. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8 A structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea.
Nat.Struct.Biol., 4, 1997
|
|
1FMC
 
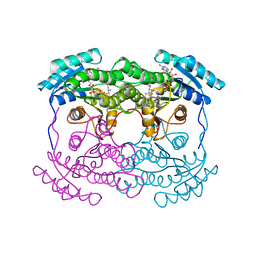 | | 7-ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEX WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1996-04-26 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
2E52
 
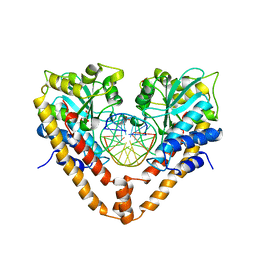 | | Crystal structural analysis of HindIII restriction endonuclease in complex with cognate DNA at 2.0 angstrom resolution | | Descriptor: | ACETATE ION, DNA (5'-D(*DGP*DCP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DGP*DGP*DC)-3'), GLYCEROL, ... | | Authors: | Watanabe, N, Sato, C, Tanaka, I. | | Deposit date: | 2006-12-18 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of restriction endonuclease HindIII in complex with its cognate DNA and divalent cations
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3A4K
 
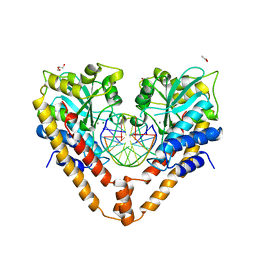 | | Crystal structural analysis of HindIII restriction endonuclease in complex with cognate DNA and divalent cations at 2.17 angstrom resolution | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*CP*CP*A)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Watanabe, N, Sato, C, Takasaki, Y, Tanaka, I. | | Deposit date: | 2009-07-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of restriction endonuclease HindIII in complex with its cognate DNA and divalent cations
Acta Crystallogr.,Sect.D, 65, 2009
|
|
8HUD
 
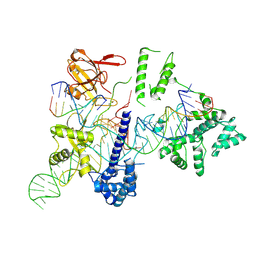 | | Cryo-EM structure of the EvCas9-sgRNA-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target DNA strand, Target DNA strand, ... | | Authors: | Tang, N, Wu, Z, Gao, Y, Chen, W, Su, M, Wang, Z, Ji, Q. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-EM structure of the EvCas9-sgRNA-target DNA ternary complex
To Be Published
|
|
5LOZ
 
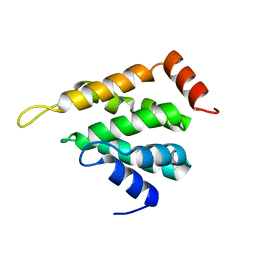 | | STRUCTURE OF YEAST ENT1 ENTH DOMAIN | | Descriptor: | Epsin-1 | | Authors: | Tanner, N, Prag, G. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A bacterial genetic selection system for ubiquitylation cascade discovery.
Nat.Methods, 13, 2016
|
|
1KOL
 
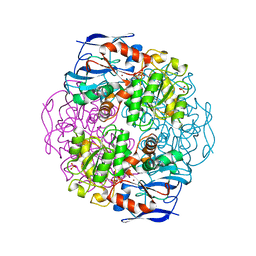 | | Crystal structure of formaldehyde dehydrogenase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ZINC ION, ... | | Authors: | Tanaka, N, Kusakabe, Y, Ito, K, Yoshimoto, T, Nakamura, K.T. | | Deposit date: | 2001-12-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Formaldehyde Dehydrogenase from Pseudomonas putida: the Structural Origin of the Tightly Bound Cofactor in Nicotinoprotein Dehydrogenases
J.mol.biol., 324, 2002
|
|
1JIQ
 
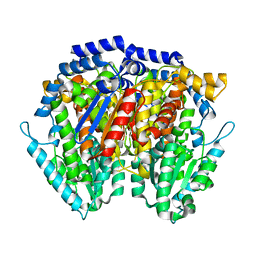 | | Crystal Structure of Human Autocrine Motility Factor | | Descriptor: | autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-07-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
1DIX
 
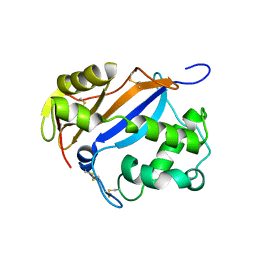 | | CRYSTAL STRUCTURE OF RNASE LE | | Descriptor: | EXTRACELLULAR RIBONUCLEASE LE | | Authors: | Tanaka, N, Nakamura, K.T. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a plant ribonuclease, RNase LE.
J.Mol.Biol., 298, 2000
|
|
1IRI
 
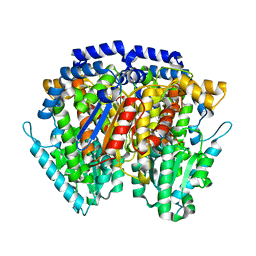 | | Crystal structure of human autocrine motility factor complexed with an inhibitor | | Descriptor: | ERYTHOSE-4-PHOSPHATE, autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-10-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
1AHH
 
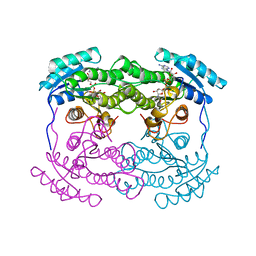 | | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-08-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
1AHI
 
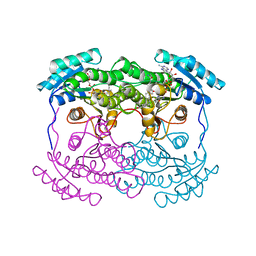 | | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEXED WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-08-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
1V8B
 
 | | Crystal structure of a hydrolase | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, adenosylhomocysteinase | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Shiraiwa, K, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-01-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of S-Adenosyl-l-Homocysteine Hydrolase from the Human Malaria Parasite Plasmodium falciparum
J.Mol.Biol., 343, 2004
|
|
1CYC
 
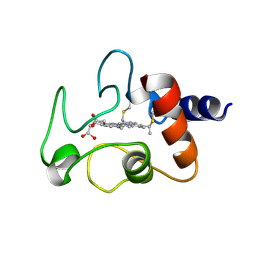 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | Descriptor: | FERROCYTOCHROME C, HEME C | | Authors: | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | Deposit date: | 1976-08-01 | | Release date: | 1976-10-06 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
1CYD
 
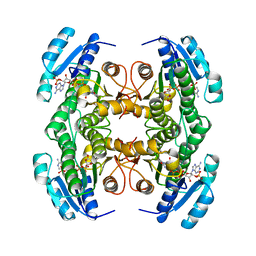 | | CARBONYL REDUCTASE COMPLEXED WITH NADPH AND 2-PROPANOL | | Descriptor: | CARBONYL REDUCTASE, ISOPROPYL ALCOHOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 A resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family.
Structure, 4, 1996
|
|
3A8U
 
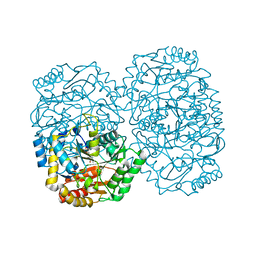 | |
7EET
 
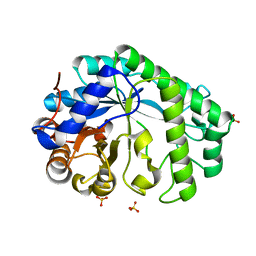 | | Mannanase KMAN from Klebsiella oxytoca KUB-CW2-3 | | Descriptor: | Mannanase KMAN, SULFATE ION | | Authors: | Pongsapipatana, N, Charoenwattanasatien, R, Pramanpol, N, Nitisinprasert, S, Keawsompong, S. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystallization, structural characterization and kinetic analysis of a GH26 beta-mannanase from Klebsiella oxytoca KUB-CW2-3.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3WQR
 
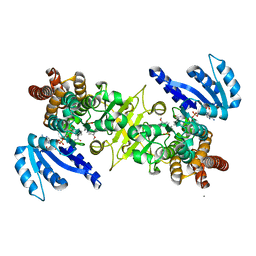 | | Crystal structure of pfdxr complexed with inhibitor-12 | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, CALCIUM ION, ... | | Authors: | Tanaka, N, Umeda, T. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding Modes of Reverse Fosmidomycin Analogs toward the Antimalarial Target IspC.
J.Med.Chem., 57, 2014
|
|
9BIZ
 
 | |
9BIY
 
 | |
9BJ0
 
 | |
6V98
 
 | | Crystal structure of Type VI secretion system effector, TseH (VCA0285) | | Descriptor: | CALCIUM ION, Cysteine hydrolase | | Authors: | Watanabe, N, Hersch, S.J, Dong, T.G, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Envelope stress responses defend against type six secretion system attacks independently of immunity proteins.
Nat Microbiol, 5, 2020
|
|
