6CA1
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | 分子名称: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
6CA3
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | 分子名称: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.743 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
6C9Z
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | 分子名称: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, Glycosyl hydrolase, family 31 | | 著者 | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | 登録日 | 2018-01-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
3B4S
 
 | |
3B4Q
 
 | |
6B6L
 
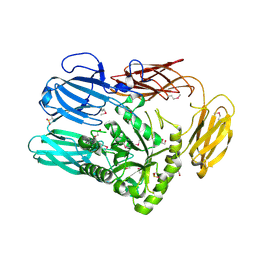 | | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, Glycosyl hydrolase family 2, ... | | 著者 | Tan, K, Joachimiak, G, Nocek, B, Enddres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-10-02 | | 公開日 | 2017-10-11 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838
To Be Published
|
|
3BJO
 
 | |
3C8G
 
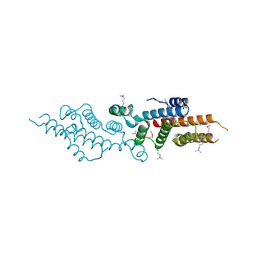 | | Crystal structure of a possible transciptional regulator YggD from Shigella flexneri 2a str. 2457T | | 分子名称: | ACETATE ION, Putative transcriptional regulator | | 著者 | Tan, K, Borovilos, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-02-12 | | 公開日 | 2008-02-19 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
3C07
 
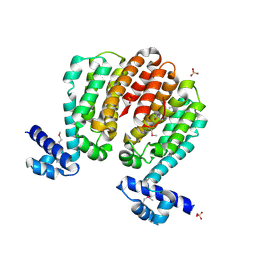 | | Crystal structure of a TetR family transcriptional regulator from Streptomyces coelicolor A3(2) | | 分子名称: | Putative tetR-family transcriptional regulator, SULFATE ION | | 著者 | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-01-18 | | 公開日 | 2008-02-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structure of a TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
To be Published
|
|
7JFQ
 
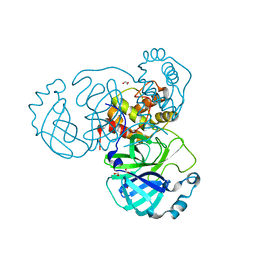 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | 著者 | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-17 | | 公開日 | 2020-07-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
1AKP
 
 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | 分子名称: | APOKEDARCIDIN | | 著者 | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | 登録日 | 1994-06-20 | | 公開日 | 1994-08-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
1BI6
 
 | |
7YML
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodobacter capsulatus | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | 著者 | Tani, K, Kanno, R, Ji, X.-C, Satoh, I, Kobayashi, Y, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Mizoguchi, A, Humbel, B.M, Madigan, M.T, Wang-Otomo, Z.-Y. | | 登録日 | 2022-07-28 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Rhodobacter capsulatus forms a compact crescent-shaped LH1-RC photocomplex.
Nat Commun, 14, 2023
|
|
4XXI
 
 | | Crystal structure of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | 分子名称: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | 著者 | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | 登録日 | 2015-01-30 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XXK
 
 | | Crystal structure of the Semet-derivative of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | 分子名称: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | 著者 | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | 登録日 | 2015-01-30 | | 公開日 | 2015-12-16 | | 最終更新日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8TV8
 
 | | Crystal structure of nontypeable Haemophilus influenzae SapA | | 分子名称: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance | | 著者 | Tanaka, K.J, Buechel, E.R, Rivera, K.G, Pinkett, H.W. | | 登録日 | 2023-08-17 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Antimicrobial Peptide Recognition Motif of the Substrate Binding Protein SapA from Nontypeable Haemophilus influenzae .
Biochemistry, 63, 2024
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | 著者 | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | 登録日 | 2023-09-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.24 Å) | | 主引用文献 | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
5SZD
 
 | | Crystal structure of Aquifex aeolicus Hfq at 1.5A | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | 著者 | Stanek, K, Patterson, J, Randolph, P.S, Mura, C. | | 登録日 | 2016-08-13 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.494 Å) | | 主引用文献 | Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5YU8
 
 | | Cofilin decorated actin filament | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Tanaka, K, Narita, A. | | 登録日 | 2017-11-21 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for cofilin binding and actin filament disassembly
Nat Commun, 9, 2018
|
|
6DQQ
 
 | |
6DQU
 
 | |
6DQR
 
 | |
3VWI
 
 | | High resolution crystal structure of FraC in the monomeric form | | 分子名称: | AMMONIUM ION, CHLORIDE ION, Fragaceatoxin C, ... | | 著者 | Tanaka, K, Morante, K, Caaveiro, J.M.M, Gonzalez-Manas, J.M, Tsumoto, K. | | 登録日 | 2012-08-23 | | 公開日 | 2013-08-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
6F9L
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose | | 分子名称: | Beta-amylase, CHLORIDE ION, alpha-D-glucopyranose-(1-4)-3-deoxy-3-fluoro-alpha-D-glucopyranose | | 著者 | Tantanarat, K, Stevenson, C.E.M, Rejzek, M, Lawson, D.M, Field, R.A. | | 登録日 | 2017-12-14 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose
To be published
|
|
6DTG
 
 | |
