7POM
 
 | |
7PP9
 
 | |
1BKJ
 
 | | NADPH:FMN OXIDOREDUCTASE FROM VIBRIO HARVEYI | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-FLAVIN OXIDOREDUCTASE, PHOSPHATE ION | | Authors: | Tanner, J.J, Lei, B, TU, S.-C, Krause, K.L. | | Deposit date: | 1998-07-08 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flavin reductase P: structure of a dimeric enzyme that reduces flavin.
Biochemistry, 35, 1996
|
|
1D49
 
 | | THE STRUCTURE OF A B-DNA DECAMER WITH A CENTRAL T-A STEP: C-G-A-T-T-A-A-T-C-G | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*AP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Grzeskowiak, K, Yanagi, K, Dickerson, R.E. | | Deposit date: | 1991-09-17 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G.
J.Mol.Biol., 225, 1992
|
|
1CER
 
 | |
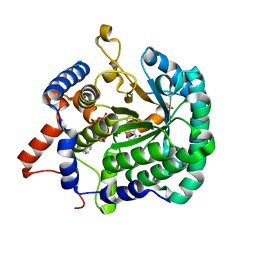
3ITG
 
 | |
6SJJ
 
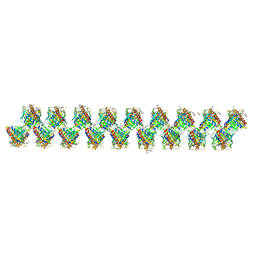 | | A new modulated crystal structure of ANS complex of St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, CITRATE ANION, ... | | Authors: | Smietanska, J, Sliwiak, J, Gilski, M, Dauter, Z, Strzalka, R, Wolny, J, Jaskolski, M. | | Deposit date: | 2019-08-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new modulated crystal structure of the ANS complex of the St John's wort Hyp-1 protein with 36 protein molecules in the asymmetric unit of the supercell.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7ST9
 
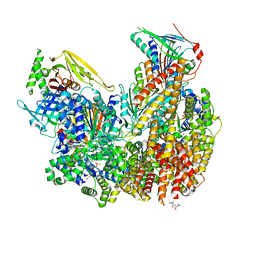 | | Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6ZS5
 
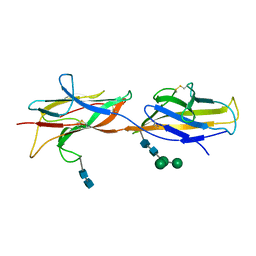 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6ZYA
 
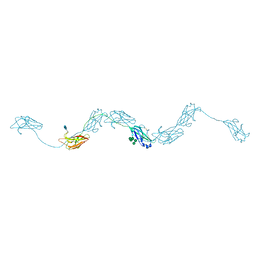 | | Extended human uromodulin filament core at 3.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
1D43
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE UP | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1D44
 
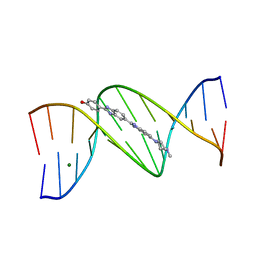 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
8CO3
 
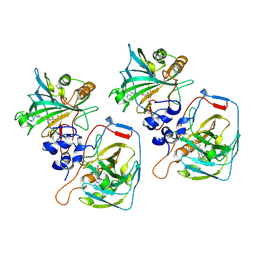 | |
8CO0
 
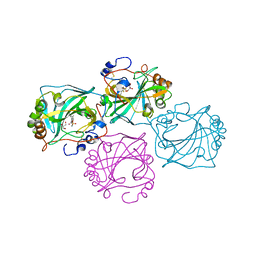 | |
7NA0
 
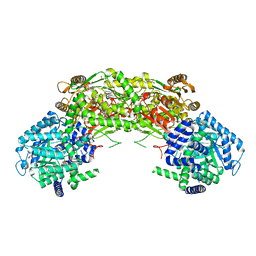 | |
7MY9
 
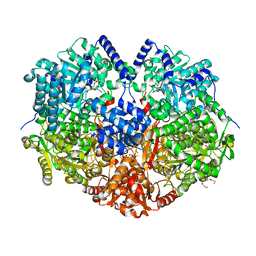 | | Structure of proline utilization A with 1,3-dithiolane-2-carboxylate bound in the proline dehydrogenase active site | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Campbell, A.C. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Photoinduced Covalent Irreversible Inactivation of Proline Dehydrogenase by S-Heterocycles.
Acs Chem.Biol., 16, 2021
|
|
7MYB
 
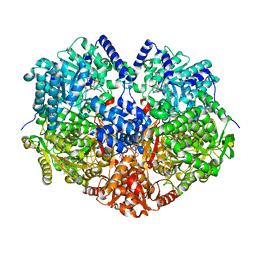 | |
7MYA
 
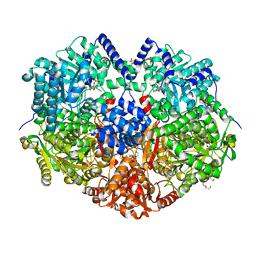 | |
7MYC
 
 | |
7STB
 
 | | Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DKQ
 
 | |
8DKP
 
 | |
8DKO
 
 | |
8Q19
 
 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with Sulfonamide | | Descriptor: | 2-but-2-ynyl-1,1,3-tris(oxidanylidene)-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
8Q18
 
 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with Sulfonamide | | Descriptor: | 1,1,3-tris(oxidanylidene)-2-(2-phenylethyl)-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
