3ZZ3
 
 | | Crystal structure of 3C protease mutant (N126Y) of coxsackievirus B3 | | 分子名称: | 3C PROTEINASE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-31 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZYE
 
 | | Crystal structure of 3C protease mutant (T68A) of coxsackievirus B3 | | 分子名称: | 3C PROTEINASE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-22 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ5
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 74 | | 分子名称: | 3C PROTEINASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-31 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ7
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 81 | | 分子名称: | 3C PROTEINASE, ETHYL (4R)-4-{[N-(TERT-BUTOXYCARBONYL)-L-PHENYLALANYL]AMINO}-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-31 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZC
 
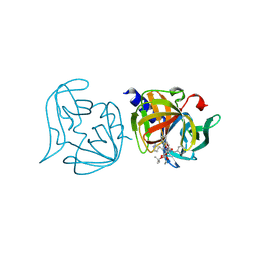 | | Crystal structure of 3C protease mutant (T68A and N126Y) of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 83 | | 分子名称: | 3C PROTEINASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-31 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ9
 
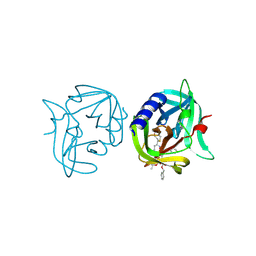 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 83 | | 分子名称: | 3C PROTEINASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-31 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ6
 
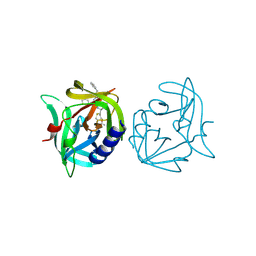 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with Michael receptor inhibitor 75 | | 分子名称: | ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE, POLYPROTEIN 3BCD | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-31 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Peptidic Ab-Nonsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
8W0K
 
 | |
8W7N
 
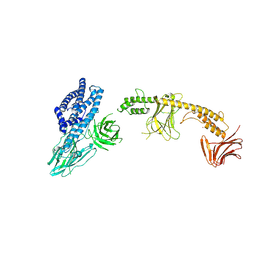 | | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis | | 分子名称: | Pesticidal crystal protein Cry1Aa, UNKNOWN ATOM OR ION | | 著者 | Tanaka, J, Abe, S, Hayakawa, T, Kojima, M, Yamashita, K, Hirata, K, Ueno, T. | | 登録日 | 2023-08-31 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 685, 2023
|
|
7STB
 
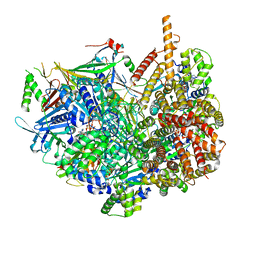 | | Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | 著者 | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | 登録日 | 2021-11-12 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ST9
 
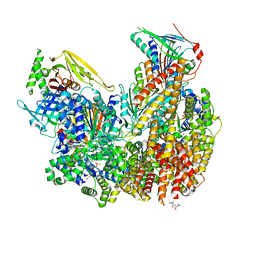 | | Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | 著者 | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | 登録日 | 2021-11-12 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7US3
 
 | |
7SQN
 
 | |
7NA0
 
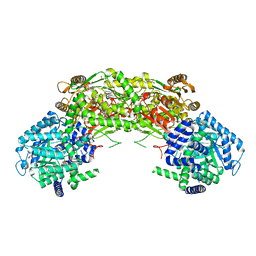 | |
7MYB
 
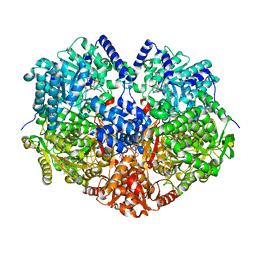 | |
7MY9
 
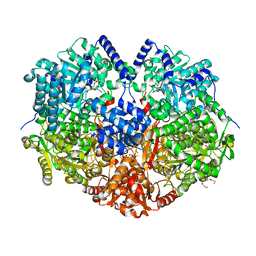 | | Structure of proline utilization A with 1,3-dithiolane-2-carboxylate bound in the proline dehydrogenase active site | | 分子名称: | 1,3-dithiolane-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tanner, J.J, Campbell, A.C. | | 登録日 | 2021-05-20 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.628 Å) | | 主引用文献 | Photoinduced Covalent Irreversible Inactivation of Proline Dehydrogenase by S-Heterocycles.
Acs Chem.Biol., 16, 2021
|
|
7MYA
 
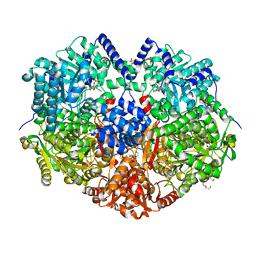 | |
7MYC
 
 | |
7STE
 
 | | Rad24-RFC ADP state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Checkpoint protein RAD24, ... | | 著者 | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | 登録日 | 2021-11-12 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | 登録日 | 2020-07-15 | | 公開日 | 2020-09-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6ZYA
 
 | | Extended human uromodulin filament core at 3.5 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | 登録日 | 2020-07-31 | | 公開日 | 2020-09-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
5KF7
 
 | |
5KF6
 
 | |
5KOW
 
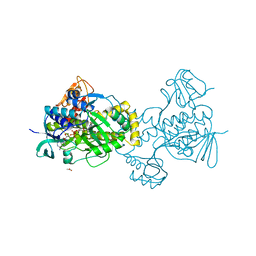 | | Structure of rifampicin monooxygenase | | 分子名称: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pentachlorophenol 4-monooxygenase | | 著者 | Tanner, J.J, Liu, L.-K. | | 登録日 | 2016-07-01 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase.
J.Biol.Chem., 291, 2016
|
|
5KOX
 
 | |
