4NBX
 
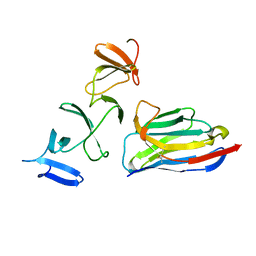 | | Crystal Structure of Clostridium difficile Toxin A fragment TcdA-A1 Bound to A20.1 VHH | | 分子名称: | A20.1 VHH, TcdA | | 著者 | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | 登録日 | 2013-10-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2014-02-12 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NBZ
 
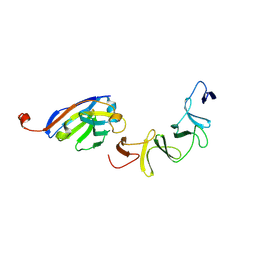 | | Crystal Structure of TcdA-A1 Bound to A26.8 VHH | | 分子名称: | A26.8 VHH, TcdA | | 著者 | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | 登録日 | 2013-10-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4FJZ
 
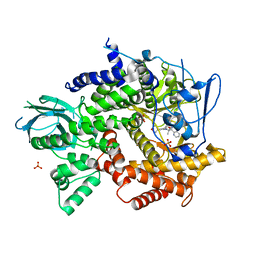 | | Crystal structure of PI3K-gamma in complex with pyrrolo-pyridine inhibitor 63 | | 分子名称: | 1'-[7-fluoro-3-methyl-2-(pyridin-2-yl)quinolin-4-yl]-6'-(morpholin-4-yl)-1',2,2',3,5,6-hexahydrospiro[pyran-4,3'-pyrrolo[3,2-b]pyridine], Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2012-06-12 | | 公開日 | 2012-10-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Discovery and in Vivo Evaluation of Dual PI3K-beta/delta inhibitors
J.Med.Chem., 55, 2012
|
|
4FLH
 
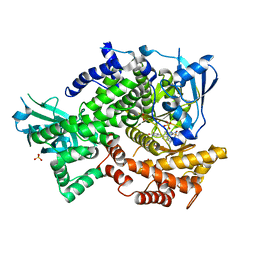 | | Crystal structure of human PI3K-gamma in complex with AMG511 | | 分子名称: | 4-(2-[(5-fluoro-6-methoxypyridin-3-yl)amino]-5-{(1R)-1-[4-(methylsulfonyl)piperazin-1-yl]ethyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2012-06-14 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Selective Class I Phosphoinositide 3-Kinase Inhibitors: Optimization of a Series of Pyridyltriazines Leading to the Identification of a Clinical Candidate, AMG 511.
J.Med.Chem., 55, 2012
|
|
4FJY
 
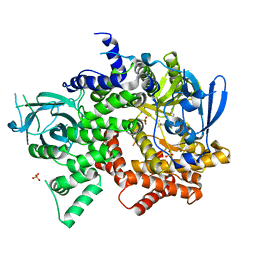 | | Crystal structure of PI3K-gamma in complex with quinoline-indoline inhibitor 24f | | 分子名称: | 4-[3,3-dimethyl-6-(morpholin-4-yl)-2,3-dihydro-1H-indol-1-yl]-7-fluoro-3-methyl-2-(pyridin-3-yl)quinoline, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2012-06-12 | | 公開日 | 2012-10-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery and in Vivo Evaluation of Dual PI3K-beta/delta inhibitors
J.Med.Chem., 55, 2012
|
|
3CS9
 
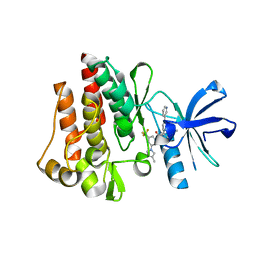 | | Human ABL kinase in complex with nilotinib | | 分子名称: | Nilotinib, Proto-oncogene tyrosine-protein kinase ABL1 | | 著者 | Cowan-Jacob, S.W, Fendrich, G, Manley, P, Liebetanz, J, Fabbro, D. | | 登録日 | 2008-04-09 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl
Cancer Cell, 7, 2005
|
|
4NBY
 
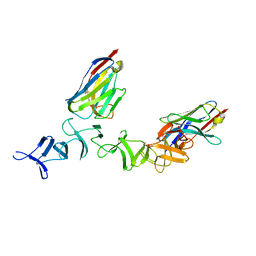 | | Crystal Structure of TcdA-A2 Bound to Two Molecules of A20.1 VHH | | 分子名称: | A20.1 VHH, Cell wall-binding repeat protein | | 著者 | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E, Klassen, J.S, Ng, K.K.S. | | 登録日 | 2013-10-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NMC
 
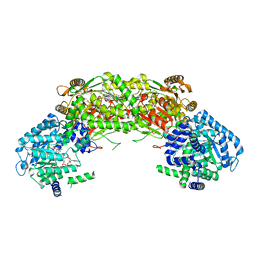 | |
4NC1
 
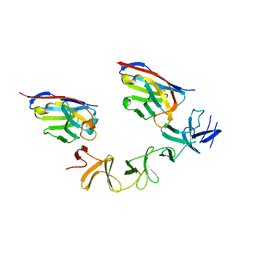 | | Crystal Structure of TcdA-A2 Bound to A20.1 VHH and A26.8 VHH | | 分子名称: | A20.1 VHH, A26.8 VHH, Cell wall-binding repeat protein | | 著者 | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | 登録日 | 2013-10-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NMD
 
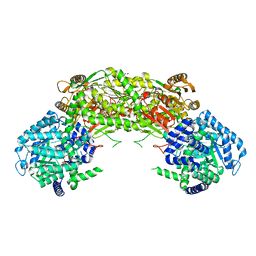 | |
4NMB
 
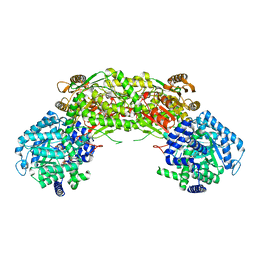 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA in complex with L-lactate | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Singh, H, Almo, S.C, Tanner, J.J. | | 登録日 | 2013-11-14 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2JMJ
 
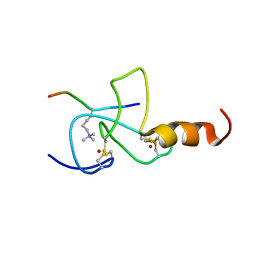 | | NMR solution structure of the PHD domain from the yeast YNG1 protein in complex with H3(1-9)K4me3 peptide | | 分子名称: | Histone H3, Protein YNG1, ZINC ION | | 著者 | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | 登録日 | 2006-11-15 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
2G94
 
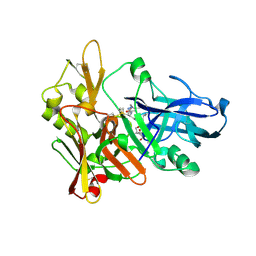 | | Crystal structure of beta-secretase bound to a potent and highly selective inhibitor. | | 分子名称: | Beta-secretase 1, N~2~-[(2R,4S,5S)-5-{[N-{[(3,5-DIMETHYL-1H-PYRAZOL-1-YL)METHOXY]CARBONYL}-3-(METHYLSULFONYL)-L-ALANYL]AMINO}-4-HYDROXY-2,7-DIMETHYLOCTANOYL]-N-ISOBUTYL-L-VALINAMIDE | | 著者 | Hong, L, Ghosh, A, Tang, J. | | 登録日 | 2006-03-05 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Design, synthesis and X-ray structure of protein-ligand complexes: important insight into selectivity of memapsin 2 (beta-secretase) inhibitors.
J.Am.Chem.Soc., 128, 2006
|
|
4NM9
 
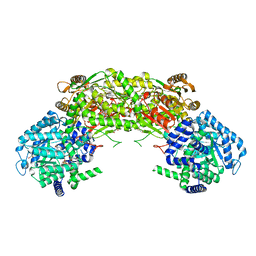 | |
4NC0
 
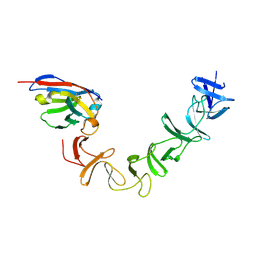 | | Crystal Structure of TcdA-A2 Bound to A26.8 VHH | | 分子名称: | A26.8 VHH, Cell wall-binding repeat protein | | 著者 | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | 登録日 | 2013-10-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
5DYS
 
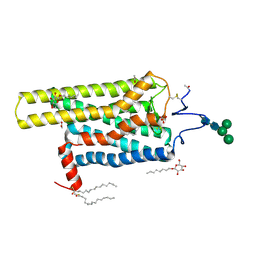 | | Crystal Structure of T94I rhodopsin mutant | | 分子名称: | ACETATE ION, PALMITIC ACID, RETINAL, ... | | 著者 | Singhal, A, Guo, Y, Matkovic, M, Schertler, G, Deupi, X, Yan, E, Standfuss, J. | | 登録日 | 2015-09-25 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural role of the T94I rhodopsin mutation in congenital stationary night blindness.
Embo Rep., 17, 2016
|
|
2JMI
 
 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | 分子名称: | Protein YNG1, ZINC ION | | 著者 | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | 登録日 | 2006-11-15 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
3CMR
 
 | | E. coli alkaline phosphatase mutant R166S in complex with phosphate | | 分子名称: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | O'Brien, P.J, Lassila, J.K, Fenn, T.D, Zalatan, J.G, Herschlag, D. | | 登録日 | 2008-03-24 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Arginine coordination in enzymatic phosphoryl transfer: evaluation of the effect of Arg166 mutations in Escherichia coli alkaline phosphatase
Biochemistry, 47, 2008
|
|
4NMA
 
 | |
4NME
 
 | |
4NC2
 
 | | Crystal structure of TcdB-B1 bound to B39 VHH | | 分子名称: | B39 VHH, Toxin B | | 著者 | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | 登録日 | 2013-10-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NMF
 
 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA inactivated by N-propargylglycine and complexed with menadione bisulfite | | 分子名称: | (2R)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, (2S)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, 1,2-ETHANEDIOL, ... | | 著者 | Singh, H, Tanner, J.J. | | 登録日 | 2013-11-14 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1XN3
 
 | | Crystal structure of Beta-secretase bound to a long inhibitor with additional upstream residues. | | 分子名称: | Beta-secretase 1, Peptidic inhibitor | | 著者 | Turner III, R.T, Hong, L, Koelsch, G, Ghosh, A.K, Tang, J. | | 登録日 | 2004-10-04 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural locations and functional roles of new subsites S5, S6, and S7 in memapsin 2 (beta-secretase).
Biochemistry, 44, 2005
|
|
4Q57
 
 | | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Song, J.-G, Kostan, J, Grishkovskaya, I, Djinovic-Carugo, K. | | 登録日 | 2014-04-16 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex
To be Published
|
|
5Y1B
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with a berberine derivative (SYSU-00679) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-3'-quinolinium propylberberine, Beta-hexosaminidase | | 著者 | Duan, Y.W, Liu, T, Zhou, Y, Tang, J.Y, Li, M, Yang, Q. | | 登録日 | 2017-07-20 | | 公開日 | 2018-01-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.207 Å) | | 主引用文献 | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
