3OIK
 
 | | Human Carbonic anhydrase II mutant A65S, N67Q (CA IX mimic) bound by 2-Ethylestradiol 3,17-O,O-bis-sulfamate | | Descriptor: | (9BETA,13ALPHA,14BETA,17ALPHA)-2-ETHYLESTRA-1(10),2,4-TRIENE-3,17-DIYL DISULFAMATE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3OKV
 
 | | Human Carbonic Anhydrase II A65S, N67Q (CA IX mimic) bound with 2-Ethylestrone 3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3P9N
 
 | | Rv2966c of M. tuberculosis is a RsmD-like methyltransferase | | Descriptor: | ACETATE ION, POSSIBLE METHYLTRANSFERASE (METHYLASE) | | Authors: | Kumar, A, Malhotra, K, Saigal, K, Sinha, K.M, Taneja, B. | | Deposit date: | 2010-10-18 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of Rv2966c protein reveals an RsmD-like methyltransferase from Mycobacterium tuberculosis and the role of its N-terminal domain in target recognition
J.Biol.Chem., 286, 2011
|
|
7CFE
 
 | | Crystal structure of RsmG methyltransferase of M. tuberculosis | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Ribosomal RNA small subunit methyltransferase G, ... | | Authors: | Bijpuria, S, Maurya, A, Kumar, P, Sharma, R, Taneja, B. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of RsmG methyltransferase of M. tuberculosis
To Be Published
|
|
1Q19
 
 | | Carbapenam Synthetase | | Descriptor: | (2S,5S)-5-CARBOXYMETHYLPROLINE, CarA, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Miller, M.T, Gerratana, B, Stapon, A, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Carbapenam Synthetase (CarA)
J.Biol.Chem., 278, 2003
|
|
1Q15
 
 | | Carbapenam Synthetase | | Descriptor: | CarA | | Authors: | Miller, M.T, Gerratana, B, Stapon, A, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Carbapenam Synthetase (CarA)
J.Biol.Chem., 278, 2003
|
|
4L69
 
 | |
4LQ6
 
 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1T8H
 
 | | 1.8 A CRYSTAL STRUCTURE OF AN UNCHARACTERIZED B. STEAROTHERMOPHILUS PROTEIN | | Descriptor: | BETA-MERCAPTOETHANOL, YlmD protein sequence homologue, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Mondragon, A, Taneja, B, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-12 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A CRYSTAL STRUCTURE OF AN UNCHARACTERIZED B. STEAROTHERMOPHILUS PROTEIN
To be Published
|
|
3DLA
 
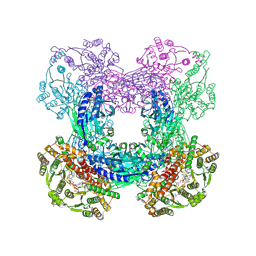 | | X-ray crystal structure of glutamine-dependent NAD+ synthetase from Mycobacterium tuberculosis bound to NaAD+ and DON | | Descriptor: | 5-OXO-L-NORLEUCINE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | LaRonde-LeBlanc, N.A, Resto, M, Gerratana, B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-03-10 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Regulation of active site coupling in glutamine-dependent NAD(+) synthetase.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3DJ6
 
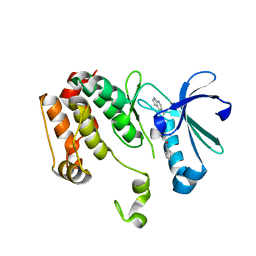 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 823. | | Descriptor: | 4-methoxy-N-phenyl-3-({3-[(1H-pyrrolo[2,3-b]pyridin-5-ylmethyl)sulfanyl]propanoyl}amino)benzamide, serine/threonine kinase 6 | | Authors: | Elling, R.A, Erlanson, D.A, Yang, W, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
3DJ5
 
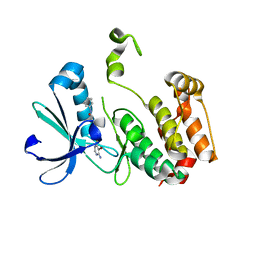 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 290. | | Descriptor: | 3-({3-[(6-amino-5-bromopyrimidin-4-yl)sulfanyl]propanoyl}amino)-4-methoxy-N-phenylbenzamide, serine/threonine kinase 6 | | Authors: | Elling, R.A, Erlanson, D.A, Yang, W, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
1D39
 
 | | COVALENT MODIFICATION OF GUANINE BASES IN DOUBLE STRANDED DNA: THE 1.2 ANGSTROMS Z-DNA STRUCTURE OF D(CGCGCG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) ION, DNA (5'-D(*CP*(CU)GP*CP*(CU)GP*CP*(CU)G)-3'), SODIUM ION | | Authors: | Kagawa, T.F, Geierstanger, B.H, Wang, A.H.-J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Covalent modification of guanine bases in double-stranded DNA. The 1.2-A Z-DNA structure of d(CGCGCG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
3DJ7
 
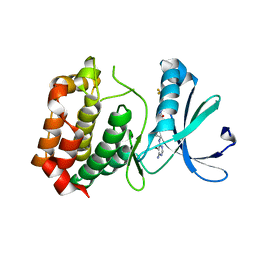 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 130. | | Descriptor: | 1-(5-{2-[(6-amino-5-bromopyrimidin-4-yl)amino]ethyl}-1,3-thiazol-2-yl)-3-[3-(trifluoromethyl)phenyl]urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Yang, W, Erlanson, D.A, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
3EPR
 
 | | Crystal structure of putative HAD superfamily hydrolase from Streptococcus agalactiae. | | Descriptor: | GLYCEROL, Hydrolase, haloacid dehalogenase-like family, ... | | Authors: | Ramagopal, U.A, Toro, R, Dickey, M, Tang, B.K, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of putative HAD superfamily hydrolase from Streptococcus agalactiae.
To be Published
|
|
7VH4
 
 | | Crystal structure of oligoribonuclease of Escherichia coli | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Badhwar, P, Taneja, B. | | Deposit date: | 2021-09-20 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of a mycobacterial oligoribonuclease reveals a unique C-terminal tail that stabilizes the homodimer.
J.Biol.Chem., 298, 2022
|
|
3SEZ
 
 | |
3SZG
 
 | | Crystal structure of C176A glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi and NaAD+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T, Gerratana, B. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
3SYT
 
 | | Crystal structure of glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi, NAD+, and glutamate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Chuenchor, W, Gerratana, B. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6511 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
3SEQ
 
 | |
1JEN
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE | | Descriptor: | PROTEIN (S-ADENOSYLMETHIONINE DECARBOXYLASE (ALPHA CHAIN)), PROTEIN (S-ADENOSYLMETHIONINE DECARBOXYLASE (BETA CHAIN)) | | Authors: | Ekstrom, J.L, Mathews, I.I, Stanley, B.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 1999-02-23 | | Release date: | 1999-06-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of human S-adenosylmethionine decarboxylase at 2.25 A resolution reveals a novel fold.
Structure Fold.Des., 7, 1999
|
|
7WIK
 
 | |
4GFJ
 
 | | Crystal structure of Topo-78, an N-terminal 78kDa fragment of topoisomerase V | | Descriptor: | GLYCEROL, Topoisomerase V, ZINC ION | | Authors: | Rajan, R, Prasad, R, Taneja, B, Wilson, S.H, Mondragon, A. | | Deposit date: | 2012-08-03 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Identification of one of the apurinic/apyrimidinic lyase active sites of topoisomerase V by structural and functional studies.
Nucleic Acids Res., 41, 2013
|
|
2L9Y
 
 | | Solution structure of the MoCVNH-LysM module from the rice blast fungus Magnaporthe oryzae protein (MGG_03307) | | Descriptor: | CVNH-LysM lectin | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Montanini, B, Kershaw, M.J, Talbot, N.J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2011-02-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-Function Analysis of a CVNH-LysM Lectin Expressed during Plant Infection by the Rice Blast Fungus Magnaporthe oryzae.
Structure, 19, 2011
|
|
3SDB
 
 | |
