1WXR
 
 | | Crystal structure of Heme Binding protein, an autotransporter hemoglobine protease from pathogenic Escherichia coli | | Descriptor: | haemoglobin protease | | Authors: | Otto, B.R, Sijbrandi, R, Luirink, J, Oudega, B, Heddle, J.G, Mizutani, K, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-01-31 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme binding protein, an autotransporter hemoglobin protease from pathogenic escherichia coli
J.Biol.Chem., 280, 2005
|
|
3G0M
 
 | | Crystal structure of cysteine desulfuration protein SufE from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Cysteine desulfuration protein sufE, ... | | Authors: | Nocek, B, Maltseva, N, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of cysteine desulfuration protein SufE from Salmonella typhimurium LT2
To be Published
|
|
3FWX
 
 | | The crystal structure of the peptide deformylase from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | Peptide deformylase, ZINC ION | | Authors: | Zhang, R, Zhou, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the peptide deformylase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3G0S
 
 | | Dihydrodipicolinate synthase from Salmonella typhimurium LT2 | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structure of dihydrodipicolinate synthase from Salmonella typhimurium LT2.
To be Published
|
|
2EX6
 
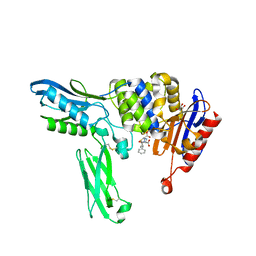 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
5UJS
 
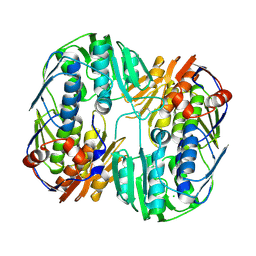 | | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.
To Be Published
|
|
1T08
 
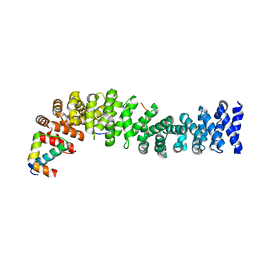 | | Crystal structure of beta-catenin/ICAT helical domain/unphosphorylated APC R3 | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin, Beta-catenin-interacting protein 1 | | Authors: | Ha, N.-C, Tonozuka, T, Stamos, J.L, Weis, W.I. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Mol.Cell, 15, 2004
|
|
5Z01
 
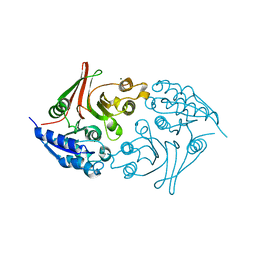 | | Native Escherichia coli L,D-carboxypeptidase A (LdcA) | | Descriptor: | MAGNESIUM ION, Murein tetrapeptide carboxypeptidase | | Authors: | Meyer, K, Tame, J.R.H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure and oligomeric form of Escherichia colil,d-carboxypeptidase A.
Biochem. Biophys. Res. Commun., 499, 2018
|
|
1UEK
 
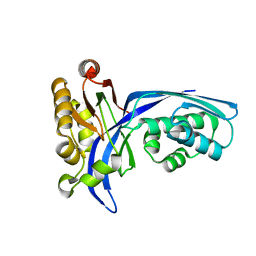 | | Crystal structure of 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | Descriptor: | 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | Authors: | Wada, T, Kuramitsu, S, Yokoyama, S, Tame, J.R.H, Park, S.Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-17 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of 4-(Cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase, an Enzyme in the Non-mevalonate Pathway of Isoprenoid Synthesis.
J.Biol.Chem., 278, 2003
|
|
3G48
 
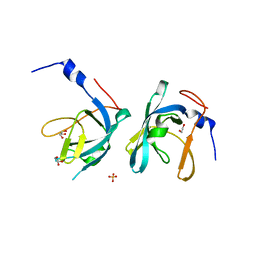 | | Crystal structure of chaperone CsaA form Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, Chaperone CsaA, GLYCEROL, ... | | Authors: | Nocek, B, Zhou, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-03 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of chaperone CsaA form Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
1RKM
 
 | | STRUCTURE OF OPPA | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
7FCI
 
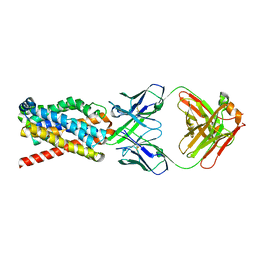 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
5US8
 
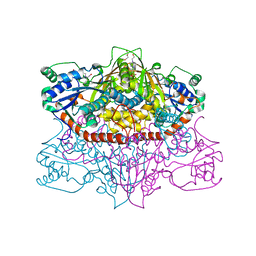 | | 2.15 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis
To Be Published
|
|
5UTU
 
 | | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD
To Be Published
|
|
5Z03
 
 | |
6AB9
 
 | | The crystal structure of the relaxed state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven cyanobacterial chloride importer in the rhodopsin family
To Be Published
|
|
1UIU
 
 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel unliganded form) | | Descriptor: | Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
1UIV
 
 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel liganded form) | | Descriptor: | NICKEL (II) ION, Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
1UIW
 
 | | Crystal Structures of Unliganded and Half-Liganded Human Hemoglobin Derivatives Cross-Linked between Lys 82beta1 and Lys 82beta2 | | Descriptor: | BUT-2-ENEDIAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Park, S.-Y, Shibayama, N, Tame, J.R.H. | | Deposit date: | 2003-07-23 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of unliganded and half-liganded human hemoglobin derivatives cross-linked between Lys 82beta1 and Lys 82beta2
Biochemistry, 43, 2004
|
|
6HUE
 
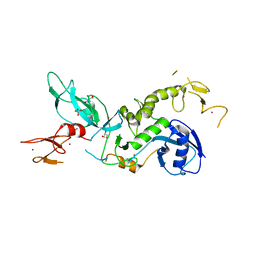 | | ParkinS65N | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | McWilliams, T.G, Barini, E, Pohjolan-Pirhonen, R, Brooks, S.P, Singh, F, Burel, S, Balk, K, Kumar, A, Montava-Garriga, L, Prescott, A.R, Hassoun, S.M, Mouton-Liger, F, Ball, G, Hills, R, Knebel, A, Ulusoy, A, Di Monte, D.A, Tamjar, J, Antico, O, Fears, K, Smith, L, Brambilla, R, Palin, E, Valori, M, Eerola-Rautio, J, Tienari, P, Corti, O, Dunnett, S.B, Ganley, I.G, Suomalainen, A, Muqit, M.M.K. | | Deposit date: | 2018-10-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Phosphorylation of Parkin at serine 65 is essential for its activation in vivo .
Open Biology, 8, 2018
|
|
6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
5F4C
 
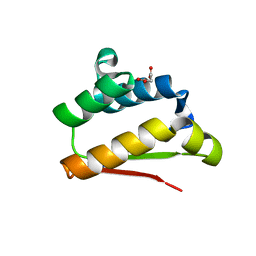 | | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium | | Descriptor: | MALONATE ION, Putative cytoplasmic protein | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium.
To Be Published
|
|
1V9F
 
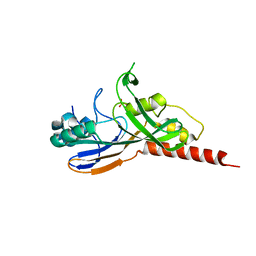 | | Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli | | Descriptor: | PHOSPHATE ION, Ribosomal large subunit pseudouridine synthase D | | Authors: | Mizutani, K, Machida, Y, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
5EVC
 
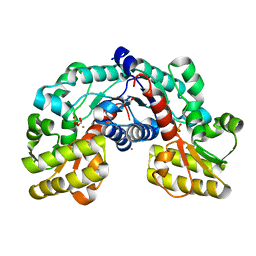 | | Crystal structure of putative aspartate racemase from Salmonella Typhimurium complexed with sulfate and potassium | | Descriptor: | CHLORIDE ION, FLUORIDE ION, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative aspartate racemase from Salmonella Typhimurium complexed with sulfate and potassium
To be published
|
|
5XG5
 
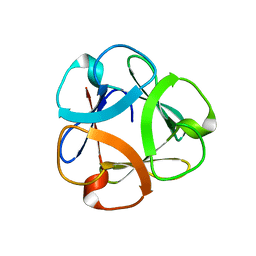 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
