8Q6T
 
 | |
8Q4G
 
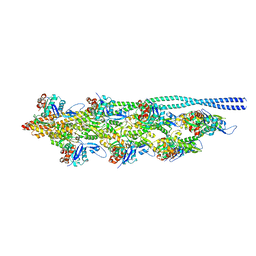 | | Thin filament from FIB milled relaxed left ventricular mouse myofibrils | | Descriptor: | Actin, alpha cardiac muscle 1, Tropomyosin alpha-1 chain | | Authors: | Tamborrini, D, Wang, Z, Wagner, T, Tacke, S, Stabrin, M, Grange, M, Kho, A.L, Bennet, P, Rees, M, Gautel, M, Raunser, S. | | Deposit date: | 2023-08-06 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of the native myosin filament in the relaxed cardiac sarcomere.
Nature, 623, 2023
|
|
1DY3
 
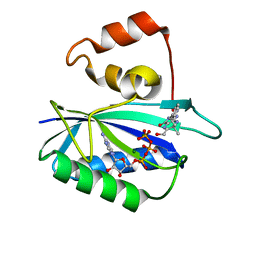 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
1K6X
 
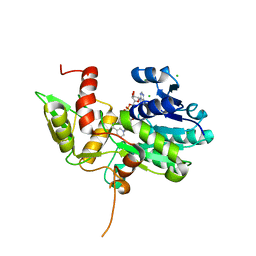 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
1K6J
 
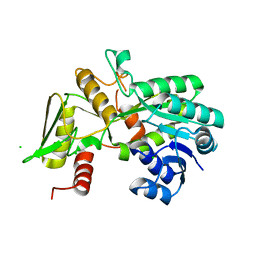 | | Crystal structure of Nmra, a negative transcriptional regulator (Monoclinic form) | | Descriptor: | CHLORIDE ION, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2001
|
|
1K6I
 
 | | Crystal structure of Nmra, a negative transcriptional regulator (Trigonal form) | | Descriptor: | CHLORIDE ION, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2001
|
|
4IFJ
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
To be Published
|
|
4IFN
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | (1R,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4IFL
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Nrf2 peptide, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4I5M
 
 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P, Neitz, R.J, Yao, N, Lin, M, Tonn, G, Zhang, H, Bova, M.P, Ren, Z, Tam, D, Ruslim, L, Baker, J, Diep, L, Fitzgerald, K, Hoffman, J, Motter, R, Fauss, D, Tanaka, P, Dappen, M, Jagodzinski, J, Chan, W, Konradi, A.W, Latimer, L, Zhu, Y.L, Artis, D.R, Sham, H.L, Anderson, J.P, Bergeron, M. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
4I11
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates. | | Descriptor: | Beta-secretase 1, N-(3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)-L-phenylalanine, ZINC ION | | Authors: | Bowers, B, Xu, Y, Yuan, S, Probst, G.D, Hom, R.K, Chan, W, Konradi, A.W, Sham, H.L, Zhu, Y.L, Beroza, P, Pan, H, Brecht, E, Yao, N, Lougheed, J, Artis, D.R, Tam, D, Bova, M. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4V5V
 
 | | Structure of respiratory syncytial virus nucleocapsid protein, P1 crystal form | | Descriptor: | RESPIRATORY SYNCYTIAL VIRUS NUCLEOCAPSID PROTEIN, RNA | | Authors: | El Omari, K, Dhaliwal, B, Ren, J, Abrescia, N.G.A, Lockyer, M, Powell, K.L, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-05-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of Respiratory Syncytial Virus Nucleocapsid Protein from Two Crystal Forms: Details of Potential Packing Interactions in the Native Helical Form.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2Q5H
 
 | | Crystal structure of apo-wildtype Glycyl-tRNA synthetase | | Descriptor: | Glycyl-tRNA synthetase | | Authors: | Cader, M.Z, Ren, J, James, P.A, Bird, L.E, Talbot, K, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy.
Febs Lett., 581, 2007
|
|
2A0J
 
 | | Crystal Structure of Nitrogen Regulatory Protein IIA-Ntr from Neisseria meningitidis | | Descriptor: | PTS system, nitrogen regulatory IIA protein | | Authors: | Ren, J, Sainsbury, S, Berrow, N.S, Alderton, D, Nettleship, J.E, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2005-06-16 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of nitrogen regulatory protein IIANtr from Neisseria meningitidis
Bmc Struct.Biol., 5, 2005
|
|
2Y7I
 
 | | Structural basis for high arginine specificity in Salmonella typhimurium periplasmic binding protein STM4351. | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Stamp, A.L, Owen, P, El Omari, K, Lockyer, M, Lamb, H.K, Charles, I.G, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-01-31 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Microcalorimetric Analyses Reveal the Structural Basis for High Arginine Specificity in the Salmonella Enterica Serovar Typhimurium Periplasmic Binding Protein Stm4351.
Proteins, 79, 2011
|
|
2XF4
 
 | | Crystal structure of Salmonella enterica serovar typhimurium YcbL | | Descriptor: | HYDROXYACYLGLUTATHIONE HYDROLASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Stamp, A, Owen, P, El Omari, K, Nichols, C, Lockyer, M, Lamb, H, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2010-05-20 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Salmonella Enterica Serovar Typhimurium Ycbl: An Unusual Type II Glyoxalase
Protein Sci., 19, 2010
|
|
4FZS
 
 | | Structure of human SNX1 BAR domain | | Descriptor: | Sorting nexin-1 | | Authors: | van Weering, J.R.T, Sessions, R.B, Traer, C.J, Kloer, D.P, Bhatia, V.K, Stamou, D, Hurley, J.H, Cullen, P.J. | | Deposit date: | 2012-07-07 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insight into vesicle-to-tubule membrane remodeling by SNX-BAR proteins
To be Published
|
|
1DS0
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE | | Descriptor: | ACETATE ION, CLAVAMINATE SYNTHASE 1, SULFATE ION | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DRT
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND PROCLAVAMINIC ACID | | Descriptor: | 2-OXOGLUTARIC ACID, 5-AMINO-3-HYDROXY-2-(2-OXO-AZETIDIN-1-YL)-PENTANOIC ACID, CLAVAMINATE SYNTHASE 1, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DS1
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II) AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DRY
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND N-ALPHA-L-ACETYL ARGININE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
2HND
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HNY
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HNZ
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with PETT-2 | | Descriptor: | 1-[2-(4-ETHOXY-3-FLUOROPYRIDIN-2-YL)ETHYL]-3-(5-METHYLPYRIDIN-2-YL)THIOUREA, PHOSPHATE ION, Reverse transcriptase/ribonuclease H | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2RCN
 
 | |
