4ZO9
 
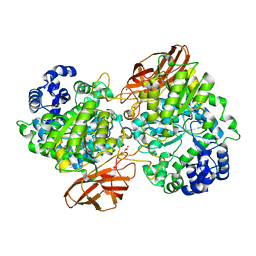 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with laminaribiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
4ZOA
 
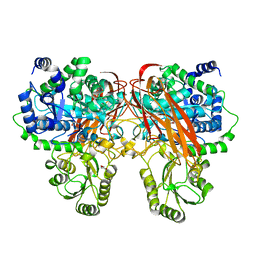 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, DI(HYDROXYETHYL)ETHER, Lin1840 protein, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
7C6A
 
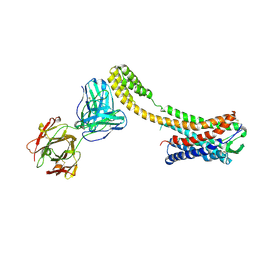 | | Crystal structure of AT2R-BRIL and SRP2070_Fab complex | | Descriptor: | IgG Light Chain, IgG heavy chain, SAR1, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
7C61
 
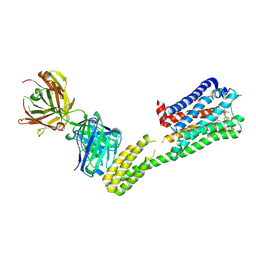 | | Crystal structure of 5-HT1B-BRIL and SRP2070_Fab complex | | Descriptor: | 5-hydroxytryptamine receptor 1B,Soluble cytochrome b562,5-hydroxytryptamine receptor 1B, Ergotamine, IGG HEAVY CHAIN, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
2Z7E
 
 | | Crystal structure of Aquifex aeolicus IscU with bound [2Fe-2S] cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, NifU-like protein, SULFATE ION | | Authors: | Shimomura, Y, Wada, K, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2007-08-20 | | Release date: | 2008-08-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The asymmetric trimeric architecture of [2Fe-2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis
J.Mol.Biol., 383, 2008
|
|
7C8M
 
 | | Crystal structure of IscU wild-type | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Nitrogen-fixing NifU domain protein | | Authors: | Kunichika, K, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of the Dimeric State of IscU Harboring Two Adjacent [2Fe-2S] Clusters Provides Mechanistic Insights into Cluster Conversion to [4Fe-4S].
Biochemistry, 60, 2021
|
|
7C8N
 
 | | Crystal structure of IscU H106A variant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Nitrogen-fixing NifU domain protein | | Authors: | Kunichika, K, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Dimeric State of IscU Harboring Two Adjacent [2Fe-2S] Clusters Provides Mechanistic Insights into Cluster Conversion to [4Fe-4S].
Biochemistry, 60, 2021
|
|
7C8O
 
 | |
7CET
 
 | | Crystal structure of D-cycloserine-bound form of cysteine desulfurase NifS from Helicobacter pylori | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEQ
 
 | |
7CER
 
 | | Crystal structure of D-cycloserine-bound form of cysteine desulfurase SufS H121A from Bacillus subtilis | | Descriptor: | Cysteine desulfurase SufS, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEO
 
 | |
7CEU
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase NifS from Helicobacter pylori | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CES
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS H121A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CNV
 
 | |
7E6A
 
 | | Crystal structure of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6E
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in D-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6C
 
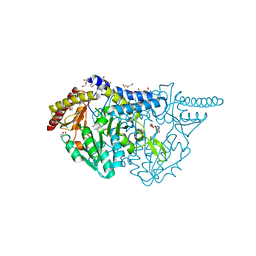 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6B
 
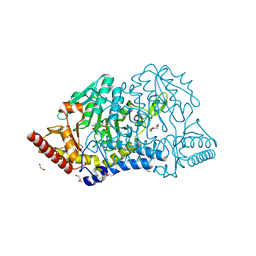 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6F
 
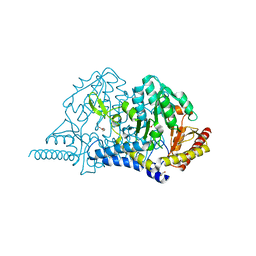 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in L-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6D
 
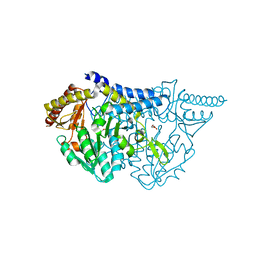 | |
6JZW
 
 | |
6JZV
 
 | | Crystal structure of SufU from Bacillus subtilis | | Descriptor: | ZINC ION, Zinc-dependent sulfurtransferase SufU | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zinc-persulfide complex for sulfur mobilization by SufU in SUF-like machinery for Fe-S cluster biosynthesis
to be published
|
|
6JO6
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Miyazaki, N, Takahashi, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-19 | | Last modified: | 2019-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
6JO5
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Miyazaki, N, Takahashi, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-19 | | Last modified: | 2019-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
