5KEF
 
 | | Structure of hypothetical Staphylococcus protein SA0856 with zinc | | Descriptor: | ACETATE ION, PhnB protein, ZINC ION | | Authors: | Battaile, K.P, Chirgadze, Y.N, Lam, R, Chan, T, Mihajlovic, V, Romanov, V, Pai, E, Mendez, V, Chirgadze, N.Y. | | Deposit date: | 2016-06-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of Staphylococcus aureus Zn-glyoxalase I: new subfamily of glyoxalase I family.
J. Biomol. Struct. Dyn., 36, 2018
|
|
4PAV
 
 | | Structure of hypothetical protein SA1046 from S. aureus. | | Descriptor: | Glyoxalase family protein | | Authors: | Battaile, K.P, Mulichak, A, Lam, R, Lam, K, Soloveychik, M, Romanov, V, Jones, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of hypothetical protein SA1046 from S. aureus.
To Be Published
|
|
4PAW
 
 | | Structure of hypothetical protein HP1257. | | Descriptor: | Orotate phosphoribosyltransferase, PHOSPHATE ION, THIOCYANATE ION, ... | | Authors: | Battaile, K.P, Lam, R, Lam, K, Romanov, V, Jones, K, Soloveychik, M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of hypothetical protein HP1257.
To Be Published
|
|
4PAU
 
 | | Hypothetical protein SA1058 from S. aureus. | | Descriptor: | CHLORIDE ION, Nitrogen regulatory protein A | | Authors: | Battaile, K.P, Wu-Brown, J, Romanov, V, Jones, K, Lam, R, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical protein SA1058 from S. aureus.
To Be Published
|
|
4P13
 
 | |
7KEY
 
 | | Protein Tyrosine Phosphatase 1B, Apo | | Descriptor: | ACETATE ION, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Battaile, K.P, Chirgadze, Y, Ruzanov, M, Romanov, V, Lam, K, Gordon, R, Lin, A, Lam, R, Pai, E, Chirgadze, N. | | Deposit date: | 2020-10-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Signal transfer in human protein tyrosine phosphatase PTP1B from allosteric inhibitor P00058.
J.Biomol.Struct.Dyn., 2021
|
|
7KLX
 
 | | Protein Tyrosine Phosphatase 1B with inhibitor | | Descriptor: | 2-(2,5-dimethyl-1H-pyrrol-1-yl)-5-hydroxybenzoic acid, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Battaile, K.P, Chirgadze, Y, Ruzanov, M, Romanov, V, Lam, K, Gordon, R, Lin, A, Lam, R, Pai, E, Chirgadze, N. | | Deposit date: | 2020-11-01 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Signal transfer in human protein tyrosine phosphatase PTP1B from allosteric inhibitor P00058.
J.Biomol.Struct.Dyn., 2021
|
|
1JQI
 
 | | Crystal Structure of Rat Short Chain Acyl-CoA Dehydrogenase Complexed With Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, short chain acyl-CoA dehydrogenase | | Authors: | Battaile, K.P, Molin-Case, J, Paschke, R, Wang, M, Bennett, D, Vockley, J, Kim, J.-J.P. | | Deposit date: | 2001-08-07 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
J.Biol.Chem., 277, 2002
|
|
2BZG
 
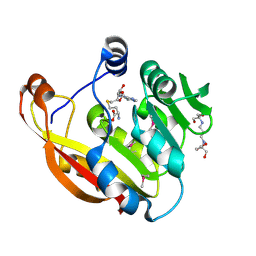 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
1RX0
 
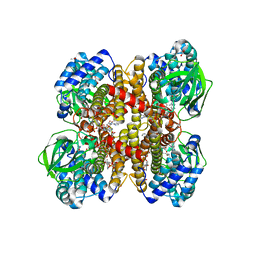 | | Crystal structure of isobutyryl-CoA dehydrogenase complexed with substrate/ligand. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acyl-CoA dehydrogenase family member 8, ... | | Authors: | Battaile, K.P, Nguyen, T.V, Vockley, J, Kim, J.J. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of Isobutyryl-CoA Dehydrogenase and Enzyme-Product Complex: COMPARISON WITH ISOVALERYL- AND SHORT-CHAIN ACYL-COA DEHYDROGENASES.
J.Biol.Chem., 279, 2004
|
|
6E6Q
 
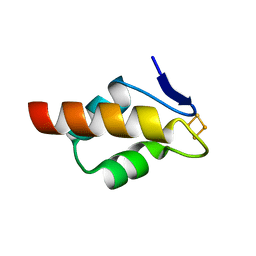 | | 1.20 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | Deposit date: | 2018-07-25 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
6E6R
 
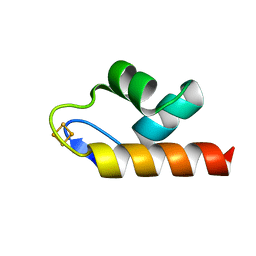 | | 1.50 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) R26E mutant from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | Deposit date: | 2018-07-25 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
2D23
 
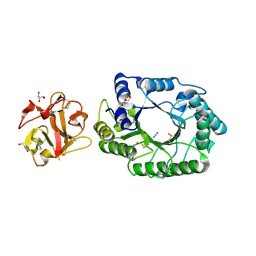 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D1Z
 
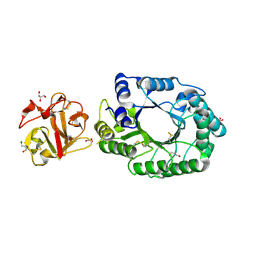 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D20
 
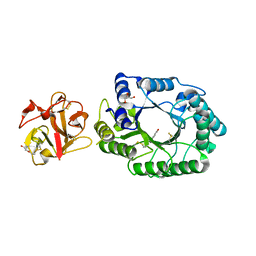 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
4LQT
 
 | | 1.10A resolution crystal structure of a superfolder green fluorescent protein (W57A) mutant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
2K1I
 
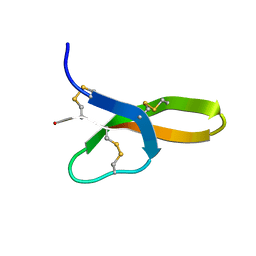 | | Synthesis, Structure and Activities of an Oral Mucosal Alpha-Defensin from Rhesus Macaque | | Descriptor: | Mucosal Alpha-Defensin | | Authors: | Vasudevan, S.V, Yuan, J, Osapay, G, Tran, P, Tai, K, Selsted, M, Cocco, M.J. | | Deposit date: | 2008-03-05 | | Release date: | 2008-10-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Synthesis, structure, and activities of an oral mucosal alpha-defensin from rhesus macaque.
J.Biol.Chem., 283, 2008
|
|
3NZZ
 
 | | Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD | | Descriptor: | Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
7T40
 
 | | Structure of MERS 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T42
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 2c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T49
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4B
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Y
 
 | | Structure of MERS 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T46
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T41
 
 | | Structure of MERS 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
