6F1H
 
 | | C1rC1s complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-galactopyranose-(1-4)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | Deposit date: | 2017-11-22 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6F1C
 
 | | C1rC1s complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1r subcomponent, ... | | Authors: | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6F39
 
 | | C1r homodimer CUB1-EGF-CUB2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1r subcomponent, ... | | Authors: | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (5.801 Å) | | Cite: | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6F1D
 
 | | CUB2 domain of C1r | | Descriptor: | CALCIUM ION, Complement C1r subcomponent, SODIUM ION | | Authors: | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2SOD
 
 | | DETERMINATION AND ANALYSIS OF THE 2 ANGSTROM STRUCTURE OF COPPER, ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Tainer, J.A, Getzoff, E.D, Richardson, J.S, Richardson, D.C. | | Deposit date: | 1980-03-25 | | Release date: | 1980-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase.
J.Mol.Biol., 160, 1982
|
|
1AKZ
 
 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
1DM5
 
 | |
2H3Q
 
 | | Solution structure of HIV-1 myrMA bound to di-C4-phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1,2-DIYL DIBUTANOATE, Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Miller, J, Tai, J, Kim, A, Ghanam, R.H, Summers, M.F. | | Deposit date: | 2006-05-22 | | Release date: | 2006-07-25 | | Last modified: | 2014-01-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3OSG
 
 | | The structure of protozoan parasite Trichomonas vaginalis Myb2 in complex with MRE-1-12 DNA | | Descriptor: | 5'-D(*AP*AP*AP*TP*AP*TP*CP*GP*TP*TP*AP*T)-3', 5'-D(*AP*TP*AP*AP*CP*GP*AP*TP*AP*TP*TP*T)-3', MYB21 | | Authors: | Jiang, I, Tsai, C.K, Chen, S.C, Wang, S.H, Amiraslanov, I, Chang, C.F, Wu, W.J, Tai, J.H, Liaw, Y.C, Huang, T.H. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 39, 2011
|
|
3OSF
 
 | | The structure of protozoan parasite Trichomonas vaginalis Myb2 in complex with MRE-2f-13 DNA | | Descriptor: | 5'-D(*CP*AP*AP*GP*AP*CP*GP*AP*TP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*AP*TP*CP*GP*TP*CP*TP*TP*G)-3', ISOPROPYL ALCOHOL, ... | | Authors: | Jiang, I, Tsai, C.K, Chen, S.C, Wang, S.H, Amiraslanov, I, Chang, C.F, Wu, W.J, Tai, J.H, Liaw, Y.C, Huang, T.H. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 39, 2011
|
|
3ZQC
 
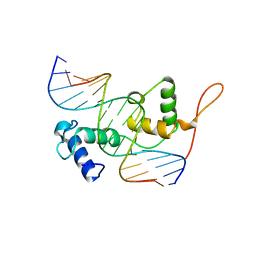 | | Structure of the Trichomonas vaginalis Myb3 DNA-binding domain bound to a promoter sequence reveals a unique C-terminal beta-hairpin conformation | | Descriptor: | MRE-1, MYB3 | | Authors: | Wei, S.-Y, Lou, Y.-C, Tsai, J.-Y, Hsu, H.-M, Tai, J.-H, Hsiao, C.-D, Chen, C. | | Deposit date: | 2011-06-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Trichomonas Vaginalis Myb3 DNA-Binding Domain Bound to a Promoter Sequence Reveals a Unique C-Terminal Beta-Hairpin Conformation.
Nucleic Acids Res., 40, 2012
|
|
1EM1
 
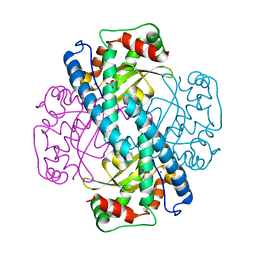 | | X-RAY CRYSTAL STRUCTURE FOR HUMAN MANGANESE SUPEROXIDE DISMUTASE, Q143A | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE, SULFATE ION | | Authors: | Leveque, V, Stroupe, M.E, Lepock, J.R, Cabelli, D.E, Tainer, J.A, Nick, H.S, Silverman, D.N. | | Deposit date: | 2000-03-14 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Multiple replacements of glutamine 143 in human manganese superoxide dismutase: effects on structure, stability, and catalysis.
Biochemistry, 39, 2000
|
|
3CRV
 
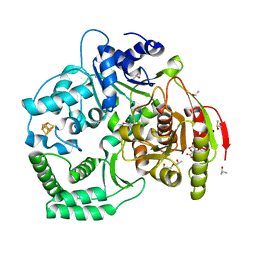 | | XPD_Helicase | | Descriptor: | CITRATE ANION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
3CRW
 
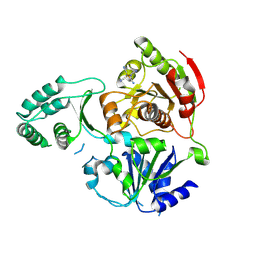 | | XPD_APO | | Descriptor: | HEXACYANOFERRATE(3-), XPD/Rad3 related DNA helicase | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
1EMH
 
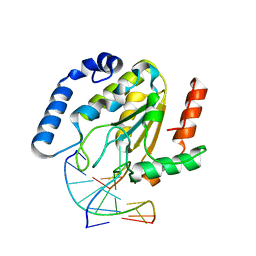 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO UNCLEAVED SUBSTRATE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(P2U)P*AP*TP*CP*TP*T)-3'), URACIL-DNA GLYCOSYLASE | | Authors: | Parikh, S.S, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EH7
 
 | |
1EMJ
 
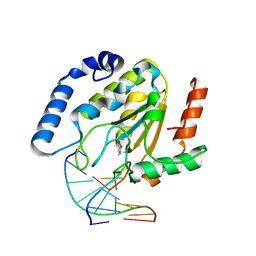 | | URACIL-DNA GLYCOSYLASE BOUND TO DNA CONTAINING A 4'-THIO-2'DEOXYURIDINE ANALOG PRODUCT | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(ASU)P*AP*TP*CP*TP*T)-3'), URACIL, ... | | Authors: | Parikh, S.S, Walcher, G, Jones, G.D, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8BYX
 
 | |
8BZM
 
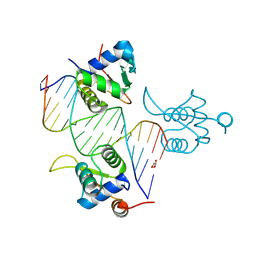 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | FOXK1-ELF1_heterodimer bound to DNA
To Be Published
|
|
2G5Z
 
 | | Structure of S65G Y66S GFP variant after spontaneous peptide hydrolysis and decarboxylation | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-02-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Understanding GFP Posttranslational Chemistry: Structures of Designed Variants that Achieve Backbone Fragmentation, Hydrolysis, and Decarboxylation.
J.Am.Chem.Soc., 128, 2006
|
|
2NV3
 
 | | Solution structure of L8A mutant of HIV-1 myristoylated matrix protein | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Loeliger, E, Luncsford, P, Liriano, M, Tai, J, Kim, A, Miller, J, Joshi, A, Freed, E.O, Summers, M.F. | | Deposit date: | 2006-11-10 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the HIV-1 matrix protein turn off the myristyl switch.
J.Mol.Biol., 366, 2007
|
|
4PPE
 
 | | human RNF4 RING domain | | Descriptor: | E3 ubiquitin-protein ligase RNF4, ZINC ION | | Authors: | Perry, J.J, Arvai, A.S, Hitomi, C, Tainer, J.A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNF4 interacts with both SUMO and nucleosomes to promote the DNA damage response.
Embo Rep., 15, 2014
|
|
2G4C
 
 | | Crystal Structure of human DNA polymerase gamma accessory subunit | | Descriptor: | DNA polymerase gamma subunit 2 | | Authors: | Fan, L, Farr, C.L, Kaguni, L.S, Tainer, J.A. | | Deposit date: | 2006-02-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase
J.Mol.Biol., 358, 2006
|
|
4P94
 
 | | The crystal structure of the soluble domain of Sulfolobus acidocaldarius FlaF (residues 35-164) | | Descriptor: | Conserved flagellar protein F, SODIUM ION | | Authors: | Tsai, C.-L, Arvai, A.S, Ishida, J.P, Tainer, J.A. | | Deposit date: | 2014-04-02 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | FlaF Is a beta-Sandwich Protein that Anchors the Archaellum in the Archaeal Cell Envelope by Binding the S-Layer Protein.
Structure, 23, 2015
|
|
2FZ4
 
 | |
