3S48
 
 | | Human Alpha-Haemoglobin Complexed with the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kumar, K.K, Jacques, D.A, Caradoc-Davies, T.T, Guss, J.M, Gell, D.A. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of alpha-haemoglobin in complex with a haemoglobin-binding domain from Staphylococcus aureus reveals the elusive alpha-haemoglobin dimerization interface
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
3SIE
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
4BJ8
 
 | | Zebavidin | | Descriptor: | BIOTIN, GLYCEROL, ZEBAVIDIN | | Authors: | Airenne, T.T, Parthiban, M, Niederhauser, B, Zmurko, J, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S. | | Deposit date: | 2013-04-17 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zebavidin
Plos One, 8, 2013
|
|
4BCS
 
 | | Crystal structure of an avidin mutant | | Descriptor: | ACETATE ION, BIOTIN, CHIMERIC AVIDIN, ... | | Authors: | Airenne, T.T, Niederhauser, B, Hytonen, V.P, Kulomaa, M.S, Johnson, M.S. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel Chimeric Avidin with Increased Thermal Stability Using DNA Shuffling.
Plos One, 9, 2014
|
|
4BN5
 
 | | Structure of human SIRT3 in complex with SRT1720 inhibitor | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, N-{2-[3-(piperazin-1-ylmethyl)imidazo[2,1-b][1,3]thiazol-6-yl]phenyl}quinoxaline-2-carboxamide, ... | | Authors: | Nguyen, G.T.T, Schaefer, S, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-05-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of Human Sirtuin 3 Complexes with Adp-Ribose and with Carba-Nad+ and Srt1720: Binding Details and Inhibition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4CAU
 
 | | THREE-DIMENSIONAL STRUCTURE OF DENGUE VIRUS SEROTYPE 1 COMPLEXED WITH 2 HMAB 14C10 FAB | | Descriptor: | ENVELOPE PROTEIN E, FAB 14C10 | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A.P, Tan, T.T, Yip, A, Schul, W, Aung, M, Kostyuchenko, V.A, Leo, Y.S, Chan, S.H, Smith, K.G, Chan, A.H, Zou, G, Ooi, E.E, Kemeny, D.M, Tan, G.K, Ng, J.K, Ng, M.L, Alonso, S, Fisher, D, Shi, P.Y, Hanson, B.J, Lok, S.M, Macary, P.A. | | Deposit date: | 2013-10-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The Structural Basis for Serotype-Specific Neutralization of Dengue Virus by a Human Antibody.
Sci.Trans.Med, 4, 2012
|
|
4C78
 
 | | Complex of human Sirt3 with Bromo-Resveratrol and ACS2 peptide | | Descriptor: | 5-[(E)-2-(4-bromophenyl)ethenyl]benzene-1,3-diol, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Sirt3 Complexes with 4'-Bromo-Resveratrol Reveal Binding Sites and Inhibition Mechanism.
Chem.Biol., 20, 2013
|
|
4BV3
 
 | | CRYSTAL STRUCTURE OF SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND NAD | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, ... | | Authors: | Gertz, M, Nguyen, N.T.T, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AYL
 
 | | Molecular structure of a metal-independent bacterial glycosyltransferase that catalyzes the synthesis of histo-blood group A antigen | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BOGT-METAL-INDEPENDENT GLYCOSYLTRANSFERASE, CALCIUM ION, ... | | Authors: | Thiyagarajan, N, Pham, T.T.K, Stinsonb, B, Sundriyala, A, Tumbale, P, Lizotte-Waniewskib, M, Brewb, K, Acharya, K.R. | | Deposit date: | 2012-06-21 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structure of a Metal-Independent Bacterial Glycosyltransferase that Catalyzes the Synthesis of Histo-Blood Group a Antigen
Sci.Rep., 2, 2012
|
|
3STQ
 
 | |
4C7B
 
 | | Complex of human Sirt3 with Bromo-Resveratrol and Fluor-De-Lys peptide | | Descriptor: | 5-[(E)-2-(4-bromophenyl)ethenyl]benzene-1,3-diol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Sirt3 Complexes with 4'-Bromo-Resveratrol Reveal Binding Sites and Inhibition Mechanism.
Chem.Biol., 20, 2013
|
|
3UQW
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, SULFATE ION, ethyl 1-{(2S,3S)-3-[(3-{[(1R)-1-(4-fluorophenyl)ethyl]carbamoyl}-5-[methyl(methylsulfonyl)amino]benzoyl)amino]-2-hydroxy-4-phenylbutyl}-1H-pyrazole-4-carboxylate | | Authors: | Chen, T.T, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility of the Flap in the Active Site of BACE1 as Revealed by Crystal Structures and MD simulations
to be published
|
|
3UQU
 
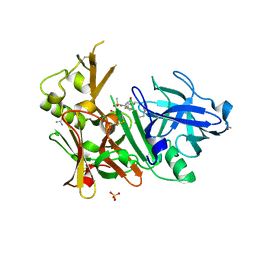 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1R)-1-(4-fluorophenyl)ethyl]-N'-[(2S,3S)-3-hydroxy-1-phenyl-4-(1H-pyrazol-1-yl)butan-2-yl]-5-[methyl(methylsulfonyl)amino]benzene-1,3-dicarboxamide, ... | | Authors: | Chen, T.T, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Flexibility of the Flap in the Active Site of BACE1 as Revealed by Crystal Structures and MD simulations
to be published
|
|
4BVG
 
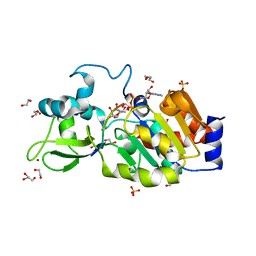 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH NATIVE ALKYLIMIDATE FORMED FROM ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
3TID
 
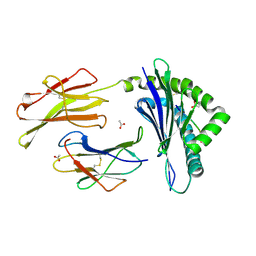 | | Crystal structure of the LCMV derived peptide GP34 in complex with the murine mhc class I H-2 Kb | | Descriptor: | ACETATE ION, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Nguyen, T.T, Shen, Z.T, Stern, L.J. | | Deposit date: | 2011-08-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Disparate epitopes mediating protective heterologous immunity to unrelated viruses share peptide-MHC structural features recognized by cross-reactive T cells.
J.Immunol., 191, 2013
|
|
3UQP
 
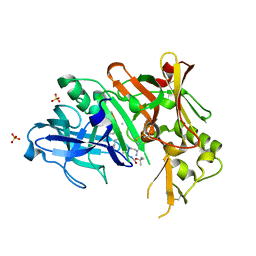 | | Crystal structure of Bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2R)-1-[(6S,9S,12S,13S,17S,20S,23R)-9-(3-AMINO-3-OXOPROPYL)-12,23-DIBENZYL-13-HYDROXY-2,2,8,20,22-PENTAMETHYL-17-(2-METHYLPROPYL)-4,7,10,15,18,21,24-HEPTAOXO-6-(PROPAN-2-YL)-3-OXA-5,8,11,16,19,22-HEXAAZATETRACOSAN-24-YL]PYRROLIDINE-2-CARBOXYLATE, SULFATE ION | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
3UQX
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1R)-1-(4-fluorophenyl)ethyl]-N'-[(2S,3S)-3-hydroxy-4-{4-[(1S)-1-hydroxyethyl]-1H-1,2,3-triazol-1-yl}-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]benzene-1,3-dicarboxamide, ... | | Authors: | Chen, T.T, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Flexibility of the Flap in the Active Site of BACE1 as Revealed by Crystal Structures and MD simulations
to be published
|
|
3UU0
 
 | | Crystal structure of L-rhamnose isomerase from Bacillus halodurans in complex with Mn | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-27 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UVA
 
 | | Crystal structure of L-rhamnose isomerase mutant W38F from Bacillus halodurans in complex with Mn | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Jeya, M, Kim, J.K, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UQR
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-15-(3-[METHYL(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-8-(2-METHYLPROPYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.056 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
3UXI
 
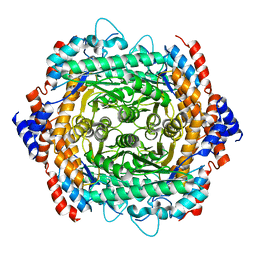 | | Crystal structure of L-rhamnose isomerase W38A mutant from Bacillus halodurans | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
4BN4
 
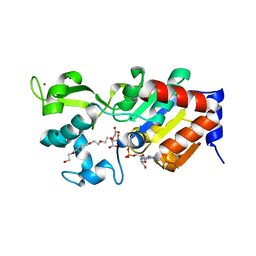 | | Structure of human SIRT3 in complex with ADP-ribose | | Descriptor: | 2-[2-[2-[2-[2-(2-hydroxyethyloxy)ethoxy]ethoxy]ethoxy]ethoxy]ethanoic acid, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-3, MITOCHONDRIAL, ... | | Authors: | Nguyen, G.T.T, Schaefer, S, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-05-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of Human Sirtuin 3 Complexes with Adp-Ribose and with Carba-Nad+ and Srt1720: Binding Details and Inhibition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3TPJ
 
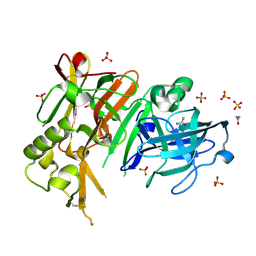 | | APO structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4BBO
 
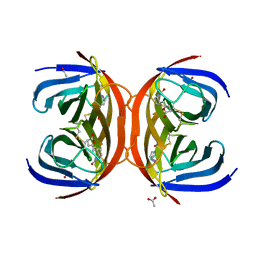 | | Crystal structure of core-bradavidin | | Descriptor: | ACETATE ION, BIOTIN, BLR5658 PROTEIN, ... | | Authors: | Airenne, T.T, Johnson, M.S, Maatta, J.A.E, Hytonen, V.H, Kulomaa, M.S. | | Deposit date: | 2012-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Core-Bradavidin
To be Published
|
|
3TPR
 
 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
