5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKA
 
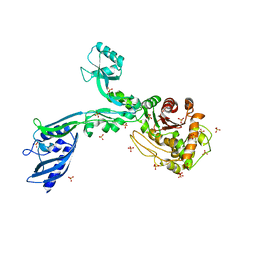 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the open conformation | | Descriptor: | SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKG
 
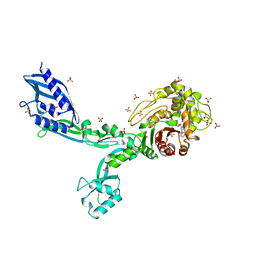 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the benzylpenicilin-bound form | | Descriptor: | OPEN FORM - PENICILLIN G, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
5SZ8
 
 | |
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
6MJ2
 
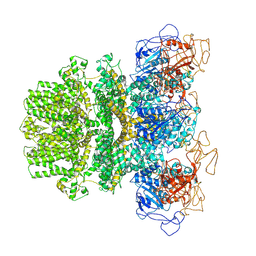 | | Human TRPM2 ion channel in a calcium- and ADPR-bound state | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-02 | | Method: | ELECTRON MICROSCOPY (6.36 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
5T3E
 
 | |
1FT5
 
 | | CRYSTAL STRUCTURE OF THE OXIDIZED STATE OF CYTOCHROME C554 FROM NITROSOMONAS EUROPAEA | | Descriptor: | CYTOCHROME C554, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Iverson, T.M, Arciero, D.M, Hooper, A.B, Rees, D.C. | | Deposit date: | 2000-09-11 | | Release date: | 2000-09-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of the oxidized and reduced states of cytochrome c554 from Nitrosomonas europaea.
J.Biol.Inorg.Chem., 6, 2001
|
|
6MIX
 
 | | Human TRPM2 ion channel in apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
1FM9
 
 | | THE 2.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE HETERODIMER OF THE HUMAN RXRALPHA AND PPARGAMMA LIGAND BINDING DOMAINS RESPECTIVELY BOUND WITH 9-CIS RETINOIC ACID AND GI262570 AND CO-ACTIVATOR PEPTIDES. | | Descriptor: | (9cis)-retinoic acid, 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA, ... | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Miller, A.B, Bledsoe, R.K, Milburn, M.V, Kliewer, S.A, Willson, T.M, Xu, H.E. | | Deposit date: | 2000-08-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors.
Mol.Cell, 5, 2000
|
|
6MKI
 
 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the ceftaroline-bound form | | Descriptor: | Ceftaroline, bound form, GLYCEROL, ... | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MIZ
 
 | |
6MKJ
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the closed conformation | | Descriptor: | penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Soares, A, D'Andrea, E.D, Jaconcic, J, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4FP1
 
 | | P. putida mandelate racemase co-crystallized with 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl) propionic acid | | Descriptor: | 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl)propanoic acid, MAGNESIUM ION, Mandelate racemase | | Authors: | Lietzan, A.D, St.Maurice, M. | | Deposit date: | 2012-06-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Potent inhibition of mandelate racemase by a fluorinated substrate-product analogue with a novel binding mode.
Biochemistry, 53, 2014
|
|
4GZ3
 
 | | Crystal structure of human O-GlcNAc Transferase with UDP and a thioglycopeptide | | Descriptor: | 2-acetamido-2-deoxy-5-thio-beta-D-glucopyranose, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
5TV1
 
 | | active arrestin-3 with inositol hexakisphosphate | | Descriptor: | Beta-arrestin-2, GLYCEROL, INOSITOL HEXAKISPHOSPHATE | | Authors: | Chen, Q, Gilbert, N.C, Perry, N.A, Vishniveteskiy, S, Gurevich, V.V, Iverson, T.M. | | Deposit date: | 2016-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of arrestin-3 activation and signaling.
Nat Commun, 8, 2017
|
|
5U35
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TRV
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
1FT6
 
 | | REDUCED STATE OF CYTOCHROME C554 FROM NITROSOMONAS EUROPAEA | | Descriptor: | CYTOCHROME C554, DITHIONITE, HEME C, ... | | Authors: | Iverson, T.M, Arciero, D.M, Hooper, A.B, Rees, D.C. | | Deposit date: | 2000-09-11 | | Release date: | 2000-09-20 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of the oxidized and reduced states of cytochrome c554 from Nitrosomonas europaea.
J.Biol.Inorg.Chem., 6, 2001
|
|
1G1A
 
 | | THE CRYSTAL STRUCTURE OF DTDP-D-GLUCOSE 4,6-DEHYDRATASE (RMLB)FROM SALMONELLA ENTERICA SEROVAR TYPHIMURIUM | | Descriptor: | DTDP-D-GLUCOSE 4,6-DEHYDRATASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Allard, S.T.M, Giraud, M.-F, Whitfield, C, Graninger, M, Messner, P, Naismith, J.H. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway.
J.Mol.Biol., 307, 2001
|
|
4GRS
 
 | | Crystal structure of a chimeric DAH7PS | | Descriptor: | Phospho-2-dehydro-3-deoxyheptonate aldolase, 2-dehydro-3-deoxyphosphoheptonate aldolase, TYROSINE | | Authors: | Cross, P.J, Allison, T.M, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2012-08-26 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering allosteric control to an unregulated enzyme by transfer of a regulatory domain
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5U89
 
 | | Crystal structure of a cross-module fragment from the dimodular NRPS DhbF | | Descriptor: | 5'-({[(2R)-3-amino-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}propyl]sulfonyl}amino)-5'-deoxyadenosine, Amino acid adenylation domain protein, MbtH domain protein | | Authors: | Tarry, M.J, Schmeing, T.M. | | Deposit date: | 2016-12-14 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | X-Ray Crystallography and Electron Microscopy of Cross- and Multi-Module Nonribosomal Peptide Synthetase Proteins Reveal a Flexible Architecture.
Structure, 25, 2017
|
|
6NOD
 
 | | Crystal structure of C. elegans PUF-8 in complex with RNA | | Descriptor: | PUF (Pumilio/FBF) domain-containing, RNA (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3') | | Authors: | Wang, Y, McCann, K.L, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
