7JRF
 
 | | CO-CO-BOUND NITROGENASE MOFE-PROTEIN FROM A. VINELANDII | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, CARBON MONOXIDE, ... | | 著者 | Spatzal, T, Perez, K.A, Buscagan, T.M, Maggiolo, A.O, Rees, D.C. | | 登録日 | 2020-08-12 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Structural Characterization of Two CO Molecules Bound to the Nitrogenase Active Site.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7K48
 
 | | Structure of NavAb/Nav1.7-VS2A chimera trapped in the resting state by tarantula toxin m3-Huwentoxin-IV | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Ion transport protein,Sodium channel protein type 9 subunit alpha chimera, Mu-theraphotoxin-Hs2a | | 著者 | Wisedchaisri, G, Tonggu, L, Gamal El-Din, T.M, McCord, E, Zheng, N, Catterall, W.A. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-02 | | 最終更新日 | 2021-01-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis for High-Affinity Trapping of the Na V 1.7 Channel in Its Resting State by Tarantula Toxin.
Mol.Cell, 81, 2021
|
|
7K3H
 
 | | Crystal structure of deep network hallucinated protein 0217 | | 分子名称: | Network hallucinated protein 0217 | | 著者 | Pellock, S.J, Anishchenko, I, Chidyausiku, T.M, Bera, A.K, DiMaio, F, Baker, D. | | 登録日 | 2020-09-11 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | De novo protein design by deep network hallucination.
Nature, 600, 2021
|
|
7JT9
 
 | |
6C1M
 
 | | NavAb NormoPP mutant | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-METHYLGUANIDINE, CHAPSO, ... | | 著者 | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | 登録日 | 2018-01-04 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.518 Å) | | 主引用文献 | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
6BSR
 
 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | 登録日 | 2017-12-04 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6P10
 
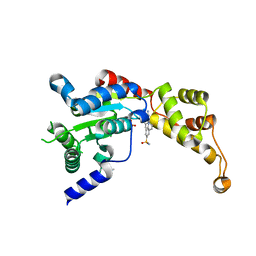 | | Structure of spastin AAA domain (N527C mutant) in complex with JNJ-7706621 inhibitor | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Drosophila melanogaster Spastin AAA domain, ... | | 著者 | Pisa, R, Cupido, T, Kapoor, T.M. | | 登録日 | 2019-05-17 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
4V5G
 
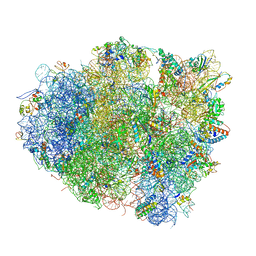 | | The crystal structure of the 70S ribosome bound to EF-Tu and tRNA | | 分子名称: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | 登録日 | 2009-09-01 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA.
Science, 326, 2009
|
|
7RZZ
 
 | |
6OZZ
 
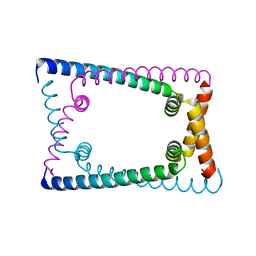 | | N terminally deleted GapR crystal structure from C. crescentus | | 分子名称: | UPF0335 protein CC_3319 | | 著者 | Tarry, M, Harmel, C, Taylor, J.A, Marczynski, G.T, Schmeing, T.M. | | 登録日 | 2019-05-16 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.304 Å) | | 主引用文献 | Structures of GapR reveal a central channel which could accommodate B-DNA.
Sci Rep, 9, 2019
|
|
3NRX
 
 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved non-motor microtubule binding protein | | 分子名称: | Protein regulator of cytokinesis 1 | | 著者 | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | 登録日 | 2010-06-30 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
4W8Q
 
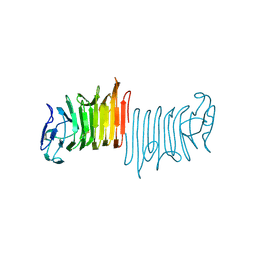 | |
3NYZ
 
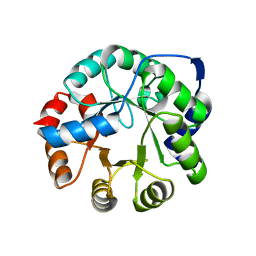 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 | | 分子名称: | Indole-3-glycerol phosphate synthase, SULFATE ION | | 著者 | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | 登録日 | 2010-07-15 | | 公開日 | 2011-06-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.514 Å) | | 主引用文献 | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NYD
 
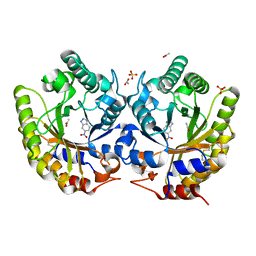 | | Crystal Structure of Kemp Eliminase HG-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | 分子名称: | 5-nitro-1H-benzotriazole, ACETATE ION, Endo-1,4-beta-xylanase, ... | | 著者 | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | 登録日 | 2010-07-14 | | 公開日 | 2011-06-29 | | 最終更新日 | 2012-04-25 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4W8R
 
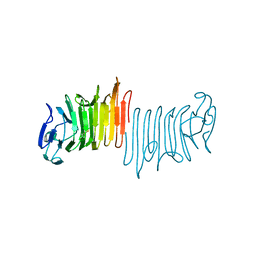 | |
3OAA
 
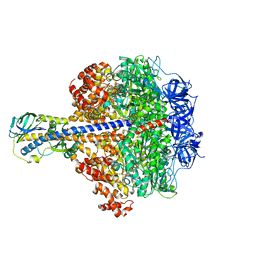 | |
4W8N
 
 | | The crystal structure of hemagglutinin from a swine influenza virus (A/swine/Missouri/2124514/2006) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Yang, H, Carney, P.J, Tumpey, T.M, Stevens, J. | | 登録日 | 2014-08-25 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Assessment of transmission, pathogenesis and adaptation of H2 subtype influenza viruses in ferrets.
Virology, 477C, 2015
|
|
6P12
 
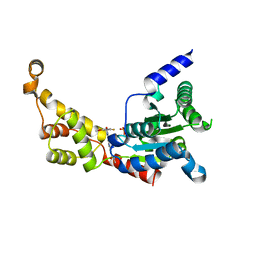 | | Structure of spastin AAA domain (wild-type) in complex with diaminotriazole-based inhibitor | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, Drosophila melanogaster Spastin AAA domain, ... | | 著者 | Pisa, R, Cupido, T, Kapoor, T.M. | | 登録日 | 2019-05-17 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.94131851 Å) | | 主引用文献 | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6P14
 
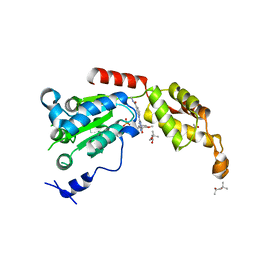 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form B) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, ACETATE ION, ... | | 著者 | Pisa, R, Cupido, T, Kapoor, T.M. | | 登録日 | 2019-05-17 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.93001127 Å) | | 主引用文献 | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
7SK4
 
 | | Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab | | 分子名称: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK6
 
 | | Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab | | 分子名称: | Atypical chemokine receptor 3, CID24 Fab heavy chain, CID24 Fab light chain, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
6P3C
 
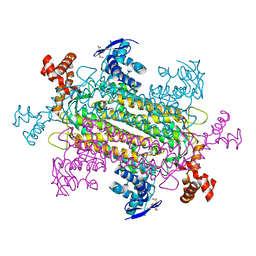 | |
7SK9
 
 | | Cryo-EM structure of human ACKR3 in complex with a small molecule partial agonist CCX662, and an intracellular Fab | | 分子名称: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK8
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab | | 分子名称: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK3
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab | | 分子名称: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | 著者 | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
