1CBU
 
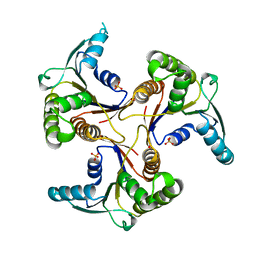 | | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE (COBU) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE, SULFATE ION | | Authors: | Thompson, T.B, Thomas, M.G, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 1998-03-12 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium determined to 2.3 A resolution,.
Biochemistry, 37, 1998
|
|
1C9K
 
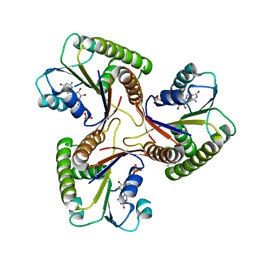 | | THE THREE DIMENSIONAL STRUCTURE OF ADENOSYLCOBINAMIDE KINASE/ ADENOSYLCOBINAMIDE PHOSPHATE GUALYLYLTRANSFERASE (COBU) COMPLEXED WITH GMP: EVIDENCE FOR A SUBSTRATE INDUCED TRANSFERASE ACTIVE SITE | | Descriptor: | ADENOSYLCOBINAMIDE KINASE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thompson, T.B, Thomas, M.G, Esclante-Semerena, J.C, Rayment, I. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase (CobU) complexed with GMP: evidence for a substrate-induced transferase active site.
Biochemistry, 38, 1999
|
|
1H9P
 
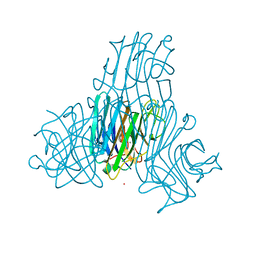 | | Crystal Structure of Dioclea guianensis Seed Lectin | | Descriptor: | CADMIUM ION, LECTIN ALPHA CHAIN, MANGANESE (II) ION | | Authors: | Romero, A, Wah, D.A, Gallego Del sol, F, Cavada, B.S, Ramos, M.V, Grangeiro, T.B, Sampaio, A.H, Calvete, J.J. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Native and Cd/Cd-Substituted Dioclea Guianensis Seed Lectin. A Novel Manganese-Binding Site and Structural Basis of Dimer-Tetramer Association
J.Mol.Biol., 310, 2001
|
|
1H9W
 
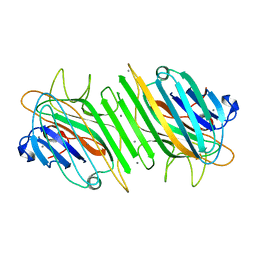 | | Native Dioclea Guianensis seed lectin | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SEED LECTIN | | Authors: | Romero, A, Wah, D.A, Sol, F.G.D, Cavada, B.S, Ramos, M.V, Grangeiro, T.B, Sampaio, A.H, Calvete, J.J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Native and Cd/Cd-Substituted Dioclea Guianensis Seed Lectin. A Novel Manganese-Binding Site and Structural Basis of Dimer-Tetramer Association
J.Mol.Biol., 310, 2001
|
|
1HDD
 
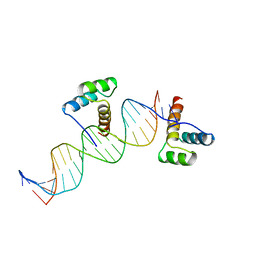 | | CRYSTAL STRUCTURE OF AN ENGRAILED HOMEODOMAIN-DNA COMPLEX AT 2.8 ANGSTROMS RESOLUTION: A FRAMEWORK FOR UNDERSTANDING HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*G P*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*C P*CP*TP*AP*A)-3'), PROTEIN (ENGRAILED HOMEODOMAIN) | | Authors: | Kissinger, C.R, Liu, B, Martin-Blanco, E, Kornberg, T.B, Pabo, C.O. | | Deposit date: | 1991-09-16 | | Release date: | 1992-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 63, 1990
|
|
1FHV
 
 | |
1JBZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT HIGH PH | | Descriptor: | 1,2-ETHANEDIOL, GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
1G5W
 
 | | SOLUTION STRUCTURE OF HUMAN HEART-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID-BINDING PROTEIN | | Authors: | Luecke, C, Rademacher, M, Zimmerman, A, van Moerkerk, H.T.B, Veerkamp, J.H, Rueterjans, H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP).
Biochem.J., 354, 2001
|
|
1JBY
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT LOW PH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
1NY1
 
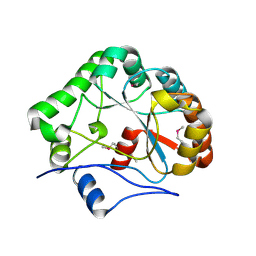 | | CRYSTAL STRUCTURE OF B. SUBTILIS POLYSACCHARIDE DEACETYLASE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR127. | | Descriptor: | Probable polysaccharide deacetylase pdaA | | Authors: | Forouhar, F, Edstrom, W, Khan, J, Ma, L, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Polysaccharide Deacetylase (PDAA_BACSU) from B. Subtilis (Pdaa_Bacsu) Northeast Structural Genomics Research Consortium (Nesg) Target Sr127
To be Published
|
|
1NNP
 
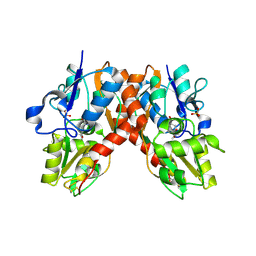 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.9 A resolution. Crystallization without zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, Glutamate receptor 2, SULFATE ION | | Authors: | Lunn, M.L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
1NNK
 
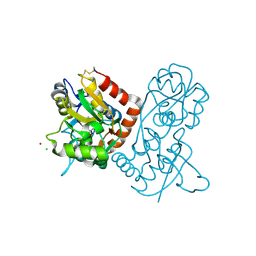 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.85 A resolution. Crystallization with zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Lunn, M.-L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
1NPD
 
 | | X-RAY STRUCTURE OF SHIKIMATE DEHYDROGENASE COMPLEXED WITH NAD+ FROM E.COLI (YDIB) NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER24 | | Descriptor: | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Kuzin, A.P, Lee, I, Rost, B, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3-A crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase
J.Biol.Chem., 278, 2003
|
|
1NXU
 
 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
1NYU
 
 | |
1N91
 
 | | Solution NMR Structure of Protein yggU from Escherichia coli. Northeast Structural Genomics Consortium Target ER14. | | Descriptor: | orf, hypothetical protein | | Authors: | Aramini, J.M, Xiao, R, Huang, Y.J, Acton, T.B, Wu, M.J, Mills, J.L, Tejero, R.T, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-11-21 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Resonance assignments for the hypothetical protein yggU from Escherichia coli.
J.Biomol.Nmr, 27, 2003
|
|
1NXZ
 
 | | X-Ray Crystal Structure of Protein yggj_haein of Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR73. | | Descriptor: | Hypothetical protein HI0303 | | Authors: | Forouhar, F, Shen, J, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional assignment based on structural analysis: crystal structure of the yggJ protein (HI0303) of Haemophilus influenzae reveals an RNA methyltransferase with a deep trefoil knot.
Proteins, 53, 2003
|
|
1M2Z
 
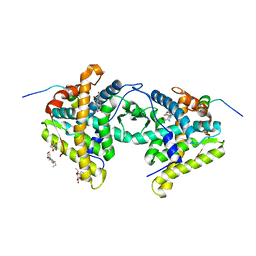 | | Crystal structure of a dimer complex of the human glucocorticoid receptor ligand-binding domain bound to dexamethasone and a TIF2 coactivator motif | | Descriptor: | DEXAMETHASONE, glucocorticoid receptor, nuclear receptor coactivator 2, ... | | Authors: | Bledsoe, R.B, Montana, V.G, Stanley, T.B, Delves, C.J, Apolito, C.J, Mckee, D.D, Consler, T.G, Parks, D.J, Stewart, E.L, Willson, T.M, Lambert, M.H, Moore, J.T, Pearce, K.H, Xu, H.E. | | Deposit date: | 2002-06-26 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel Mode of Receptor Dimerization and Coactivator Recognition
Cell(Cambridge,Mass.), 110, 2002
|
|
1MZG
 
 | | X-Ray Structure of SufE from E.coli Northeast Structural Genomics (NESG) Consortium Target ER30 | | Descriptor: | SufE Protein | | Authors: | Kuzin, A, Edstrom, W.C, Xiao, R, Acton, T.B, Rost, B, Tong, L, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes
J.Mol.Biol., 344, 2004
|
|
1NY8
 
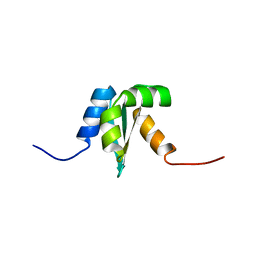 | | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115 | | Descriptor: | Protein yrbA | | Authors: | Swapna, G.V.T, Huang, J.Y, Acton, T.B, Shastry, R, Chiang, Y.-W, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115
To be Published
|
|
1NY4
 
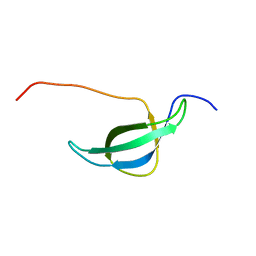 | | Solution structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii. Northeast Structural Genomics Consortium target JR19. | | Descriptor: | 30S ribosomal protein S28E | | Authors: | Aramini, J.M, Cort, J.R, Huang, Y.J, Xiao, R, Acton, T.B, Ho, C.K, Shih, L.-Y, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii.
Protein Sci., 12, 2003
|
|
1NYS
 
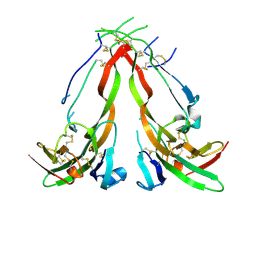 | |
1OY1
 
 | | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105 | | Descriptor: | PUTATIVE sigma cross-reacting protein 27A | | Authors: | Benach, J, Edstrom, W, Ma, L.C, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-03 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105
To be Published
|
|
1ON0
 
 | | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR144 | | Descriptor: | CHLORIDE ION, SULFATE ION, YycN protein | | Authors: | Forouhar, F, Shen, J, Kuzin, A, Chiang, Y, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-26 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis
To be Published
|
|
2K5E
 
 | | SOLUTION STRUCTURE OF PUTATIVE UNCHARACTERIZED PROTEIN GSU1278 FROM METHANOCALDOCOCCUS JANNASCHII, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET GsR195 | | Descriptor: | uncharacterized protein | | Authors: | Liu, G, Zhao, L, Ciccosanti, C, Jiang, M, Xiao, R, Swapna, G, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF PUTATIVE UNCHARACTERIZED PROTEIN GSU1278 FROM METHANOCALDOCOCCUS JANNASCHII, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET GsR195
To be Published
|
|
