3TP4
 
 | | Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | Descriptor: | De Novo design cysteine esterase FR29 | | Authors: | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U4T
 
 | | Crystal Structure of the C-terminal part of the TPR repeat-containing protein Q11TI6_CYTH3 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target ChR11B. | | Descriptor: | TPR repeat-containing protein | | Authors: | Vorobiev, S, Neely, H, Chen, Y, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-10 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Crystal Structure of the C-terminal part of the TPR repeat-containing protein Q11TI6_CYTH3 from Cytophaga hutchinsonii.
To be Published
|
|
3SYX
 
 | | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein. Northeast Structural Genomics Consortium Target HR5538B. | | Descriptor: | Sprouty-related, EVH1 domain-containing protein 1, YTTRIUM (III) ION | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein.
To be Published
|
|
3SM3
 
 | | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR262. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, SAM-dependent methyltransferases | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Snyder, A, Patel, P, Xiao, R, Ciccosanti, C, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei.
To be Published, 2009
|
|
6GX3
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 1 | | Descriptor: | 4-chloranyl-~{N}-oxidanyl-1-benzothiophene-2-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
3SH3
 
 | | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii STANDL | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rangel, T.B.A, Rocha, B.A.M, Bezerra, G.A, Bezerra, M.J.B, Nascimento, K.S, Nagano, C.S, Sampaio, A.H, Assreuy, A.M.S, Gruber, K, Delatorre, P, Cavada, B.S. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii Standl.
Biochimie, 94, 2012
|
|
3SLO
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023D mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
3SXW
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69. | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Patel, P, Xiao, R, Maglaqui, M, Ciccosanti, C, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69.
To be Published
|
|
3S84
 
 | | Dimeric apoA-IV | | Descriptor: | Apolipoprotein A-IV, SULFATE ION | | Authors: | Deng, X, Davidson, W.S, Thompson, T.B. | | Deposit date: | 2011-05-27 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Dimeric Apolipoprotein A-IV and Its Mechanism of Self-Association.
Structure, 20, 2012
|
|
3SLJ
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
6HDD
 
 | |
3SWV
 
 | | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A | | Descriptor: | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-14 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BrefeldinA-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A
To be Published
|
|
6HQY
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with PCI-34051 | | Descriptor: | 1-[(4-methoxyphenyl)methyl]-~{N}-oxidanyl-indole-6-carboxamide, GLYCEROL, Histone deacetylase, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTG
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 4 | | Descriptor: | 3-benzamido-4-chloranyl-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTZ
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 8 | | Descriptor: | 4-methoxy-~{N}-oxidanyl-3-(2-phenylethanoylamino)benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
3SEK
 
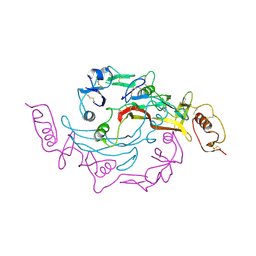 | | Crystal Structure of the Myostatin:Follistatin-like 3 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Follistatin-related protein 3, Growth/differentiation factor 8 | | Authors: | Cash, J.N, Thompson, T.B. | | Deposit date: | 2011-06-10 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of myostatinfollistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding.
J.Biol.Chem., 287, 2012
|
|
3SLT
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023S Mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-25 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
6GXU
 
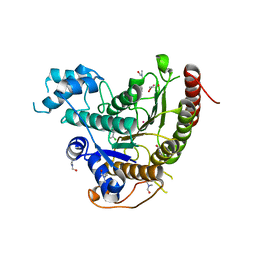 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 3 | | Descriptor: | (~{E})-3-[2-(4-chlorophenyl)sulfanylphenyl]-~{N}-oxidanyl-prop-2-enamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
6GXW
 
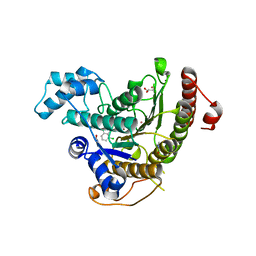 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 4 | | Descriptor: | (~{E})-3-[2-[[2,6-bis(chloranyl)phenyl]methoxy]phenyl]-~{N}-oxidanyl-prop-2-enamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
3TC7
 
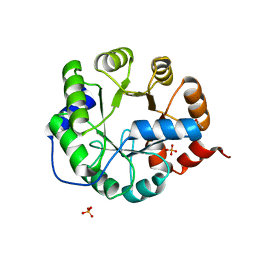 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
6HSG
 
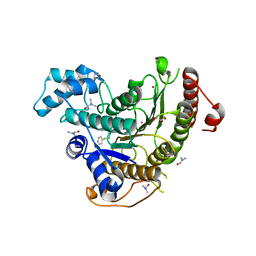 | | Crystal structure of Schistosoma mansoni HDAC8 H292M mutant complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HU1
 
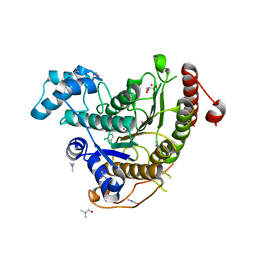 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 10 | | Descriptor: | 4-chloranyl-3-[(2,4-dichlorophenyl)carbonylamino]-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HU0
 
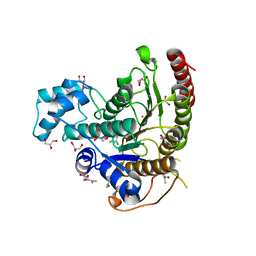 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 9 | | Descriptor: | 3-[(2,4-dichlorophenyl)carbonylamino]-4-methoxy-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HYE
 
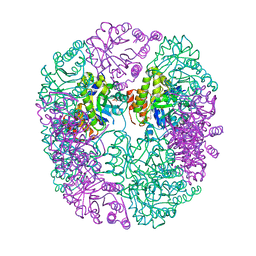 | | PDX1.2/PDX1.3 complex (PDX1.3:K97A) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
