8E53
 
 | | MicroED structure of proteinase K recorded on K3 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8E54
 
 | | MicroED structure of triclinic lysozyme recorded on K3 | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
5DD0
 
 | | Crystal structures in an anti-HIV antibody lineage from immunization of Rhesus macaques | | Descriptor: | ANTI-HIV ANTIBODY DH570 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY DH570 FAB HEAVY LIGHT, oligo peptide | | Authors: | Zhang, R, Verkoczy, L, Wiehe, K, Alam, S.M, Nicely, N.I, Santra, S, Bradley, T, Pemble, C, Gao, F, Montefiori, D.C, Bouton-Verville, H, Kelsoe, G, Parks, R, Foulger, A, Tomaras, G, Keple, T.B, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages.
Sci Transl Med, 8, 2016
|
|
5DD1
 
 | | Crystal structures in an anti-HIV antibody lineage from immunization of Rhesus macaques | | Descriptor: | ANTI-HIV ANTIBODY DH570 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY DH570 FAB LIGHT CHAIN | | Authors: | Zhang, R, Verkoczy, L, Wiehe, K, Alam, S.M, Nicely, N.I, Santra, S, Bradley, T, Pemble, C, Gao, F, Montefiori, D.C, Bouton-Verville, H, Kelsoe, G, Parks, R, Foulger, A, Tomaras, G, Keple, T.B, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages.
Sci Transl Med, 8, 2016
|
|
7Z7D
 
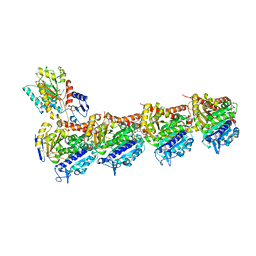 | | Tubulin-Todalam-Vinblastine-complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Roy, B, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Z5S
 
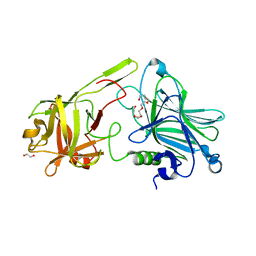 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin, HEXAETHYLENE GLYCOL, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
7Z5T
 
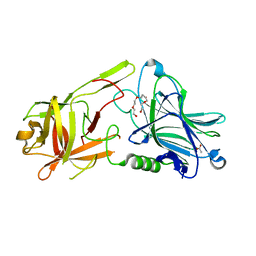 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain | | Descriptor: | ACETATE ION, Botulinum neurotoxin, PENTAETHYLENE GLYCOL | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
5GPO
 
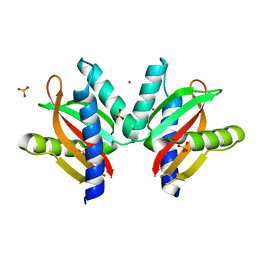 | | The sensor domain structure of the zinc-responsive histidine kinase CzcS from Pseudomonas Aeruginosa | | Descriptor: | SULFATE ION, Sensor protein CzcS, ZINC ION | | Authors: | Wang, D, Chen, W.Z, Huang, S.Q, Liu, X.C, Hu, Q.Y, Wei, T.B, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa
PLoS Pathog., 13, 2017
|
|
5GAJ
 
 | | Solution NMR structure of De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258 | | Descriptor: | DE NOVO DESIGNED PROTEIN OR258 | | Authors: | Liu, G, Castelllanos, J, Koga, R, Koga, N, Xiao, R, Pederson, K, Janjua, H, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258
To Be Published
|
|
5HK5
 
 | |
6Y93
 
 | | Crystal structure of the DNA-binding domain of the Nucleoid Occlusion Factor (Noc) complexed to the Noc-binding site (NBS) | | Descriptor: | Noc Binding Site (NBS), Nucleoid occlusion protein | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Chan, E, Lo, R, Tan, X, Noy, A, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
6YMB
 
 | | MicroED structure of human carbonic anhydrase II | | Descriptor: | ZINC ION, carbonic anhydrase 2 | | Authors: | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
6YMA
 
 | | MicroED structure of acetazolamide-bound human carbonic anhydrase II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
6Z1Y
 
 | | Crystal structure of type-I ribosome-inactivating protein trichobakin (TBK) | | Descriptor: | SODIUM ION, Trichobakin | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Britikov, V.V, Bocharov, E.V, Britikova, E.V, Le, T.B.T, Phan, C.V, Popov, V.O, Usanov, S.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of type-I ribosome-inactivating protein trichobakin (TBK)
To Be Published
|
|
5J9C
 
 | | Crystal structure of peroxiredoxin Asp f3 C31S/C61S variant | | Descriptor: | MAGNESIUM ION, peroxiredoxin Asp f3 | | Authors: | Bzymek, K.P, Williams, J.C, Hong, T.B, Bagramyan, K, Kalkum, M. | | Deposit date: | 2016-04-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | The Crystal Structure of Peroxiredoxin Asp f3 Provides Mechanistic Insight into Oxidative Stress Resistance and Virulence of Aspergillus fumigatus.
Sci Rep, 6, 2016
|
|
1TTZ
 
 | | X-ray structure of Northeast Structural Genomics target protein XcR50 from X. campestris | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Ho, C.K, Cooper, B, Ma, L.-C, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-ray structure of Northeast Structural Genomics target protein XcR50 from X. campestris
To be Published
|
|
1TUW
 
 | | Structural and Functional Analysis of Tetracenomycin F2 Cyclase from Streptomyces glaucescens: A Type-II Polyketide Cyclase | | Descriptor: | SULFATE ION, Tetracenomycin polyketide synthesis protein tcmI | | Authors: | Thompson, T.B, Katayama, K, Watanabe, K, Hutchinson, C.R, Rayment, I. | | Deposit date: | 2004-06-25 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens. A type II polyketide cyclase.
J.Biol.Chem., 279, 2004
|
|
1TZA
 
 | | X-ray structure of Northeast Structural Genomics Consortium target SoR45 | | Descriptor: | CACODYLATE ION, apaG protein | | Authors: | Kuzin, A.P, Vorobiev, S.M, Edstrom, W, Acton, T.B, Shastry, R, Ma, L.-C, Cooper, B, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of Northeast Structural Genomics Consortium
target SoR45
To be Published
|
|
1TVG
 
 | | X-ray structure of human PP25 gene product, HSPC034. Northeast Structural Genomics Target HR1958. | | Descriptor: | CALCIUM ION, LOC51668 protein, SAMARIUM (III) ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
1TWD
 
 | | Crystal Structure of the Putative Copper Homeostasis Protein (CutC) from Shigella flexneri, Northeast Structural Genomics Target SfR33 | | Descriptor: | Copper homeostasis protein cutC | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Ma, L.-C, Shastry, R, Conover, K, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-30 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Putative Copper Homeostasis Protein (CutC) from Shigella flexneri, Northeast Structural Genomics Target SfR33
To be Published
|
|
7P3S
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 12 | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Shaik, T.B, Romier, C. | | Deposit date: | 2021-07-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Synthesis, structure-activity relationships, cocrystallization and cellular characterization of novel smHDAC8 inhibitors for the treatment of schistosomiasis.
Eur.J.Med.Chem., 225, 2021
|
|
7OL9
 
 | |
7OLG
 
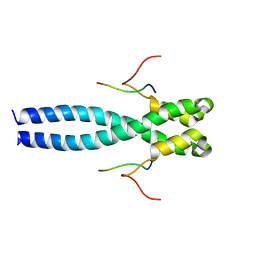 | | EB1 bound to MACF peptide | | Descriptor: | 11MACF, Microtubule-associated protein RP/EB family member 1 | | Authors: | Almeida, T.B, Barsukov, I.L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | EB1 bound to MACF peptide
To Be Published
|
|
7PBJ
 
 | |
7PBX
 
 | |
