1HUJ
 
 | |
5Y7I
 
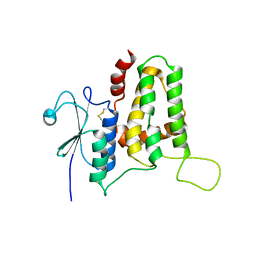 | | Structure of tilapia fish CLIC2 | | Descriptor: | chloride intracellular channel protein 2 | | Authors: | Swaminathan, K, Zeng, J. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tilapia and human CLIC2 structures are highly conserved.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
1HUK
 
 | |
1ZME
 
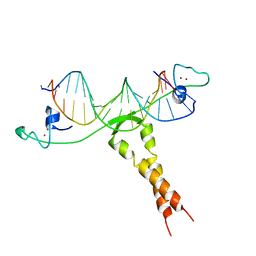 | | CRYSTAL STRUCTURE OF PUT3/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*AP*GP*(5IU)P*TP*GP*GP*CP*TP*(5IU)P*CP*CP*CP*G)-3'), DNA (5'-D(*AP*CP*GP*GP*GP*AP*AP*GP*CP*CP*AP*AP*CP*TP*CP*CP*G)-3'), PROLINE UTILIZATION TRANSCRIPTION ACTIVATOR, ... | | Authors: | Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-08-06 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
3TVD
 
 | | Crystal Structure of Mouse RhoA-GTP complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Swaminathan, K, Pal, K, Jobichen, C. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Crystal structure of mouse RhoA:GTPgammaS complex in a centered lattice.
J.Struct.Funct.Genom., 13, 2012
|
|
3PLQ
 
 | | Crystal structure of PKA type I regulatory subunit bound with Rp-8-Br-cAMPS | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-(6-amino-8-bromo-9H-purin-9-yl)tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-sulfide, ZINC ION, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Swaminathan, K. | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: Conformational selection highlights a new concept in allosteric inhibitor design
To be Published
|
|
4V0R
 
 | | DENGUE VIRUS FULL LENGTH NS5 COMPLEXED WITH GTP AND SAH | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
6LKB
 
 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
5GIM
 
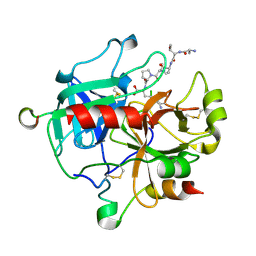 | | Crystal structure of thrombin-avathrin complex | | Descriptor: | C-terminal peptide from Putative uncharacterized protein avahiru, N-terminal peptide from Putative uncharacterized protein avahiru, Thrombin light chain, ... | | Authors: | Kini, R.M, Koh, C.Y, Iyer, J.K, Swaminathan, K. | | Deposit date: | 2016-06-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Avathrin: a novel thrombin inhibitor derived from a multicopy precursor in the salivary glands of the ixodid tick, Amblyomma variegatum.
FASEB J., 31, 2017
|
|
7W7R
 
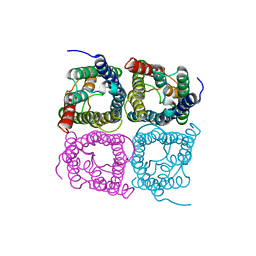 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7W7S
 
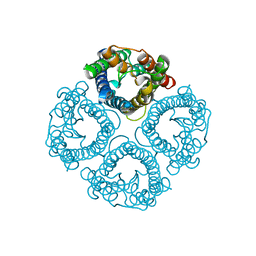 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
5DTO
 
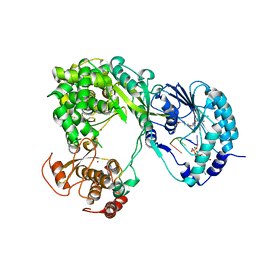 | | Dengue virus full length NS5 complexed with viral Cap 0-RNA and SAH | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Soh, T.S, Lim, S.P, Chung, K.Y, Swaminathan, K, Vasudevan, S.G, Shi, P.-Y, Lescar, J, Luo, D. | | Deposit date: | 2015-09-18 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Molecular basis for specific viral RNA recognition and 2'-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5)
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3B23
 
 | | Crystal structure of thrombin-variegin complex: Insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors | | Descriptor: | Thrombin heavy chain, Thrombin light chain, Variegin | | Authors: | Koh, C.Y, Kumar, S, Swaminathan, K, Kini, R.M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with s-variegin: insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors
Plos One, 6, 2011
|
|
4V0Q
 
 | | Dengue Virus Full Length NS5 Complexed with SAH | | Descriptor: | ACETATE ION, GLYCEROL, NS5 POLYMERASE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
7C4O
 
 | | Solution structure of the Orange domain from human protein HES1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Fan, J.S, Nayak, A, Swaminathan, K. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Induction of Transcriptional Inhibitor Hairy and Enhancer of Split Homolog-1 and the Related Repression of Tumor-Suppressor Thioredoxin-Interacting Protein Are Important Components of Cell-Transformation Program Imposed by Oncogenic Kinase Nucleophosmin-Anaplastic Lymphoma Kinase.
Am J Pathol, 2022
|
|
4URS
 
 | | Crystal Structure of GGDEF domain from T.maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE, ... | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
3K1D
 
 | | Crystal structure of glycogen branching enzyme synonym: 1,4-alpha-D-glucan:1,4-alpha-D-GLUCAN 6-glucosyl-transferase from mycobacterium tuberculosis H37RV | | Descriptor: | 1,4-alpha-glucan-branching enzyme | | Authors: | Pal, K, Kumar, S, Swaminathan, K. | | Deposit date: | 2009-09-27 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of full-length Mycobacterium tuberculosis H37Rv glycogen branching enzyme: insights of N-terminal beta-sandwich in substrate specificity and enzymatic activity
J.Biol.Chem., 285, 2010
|
|
1KHI
 
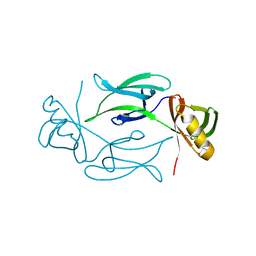 | | CRYSTAL STRUCTURE OF HEX1 | | Descriptor: | Hex1 | | Authors: | Yuan, P, Swaminathan, K. | | Deposit date: | 2001-11-30 | | Release date: | 2002-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa
NAT.STRUCT.BIOL., 10, 2003
|
|
3BC9
 
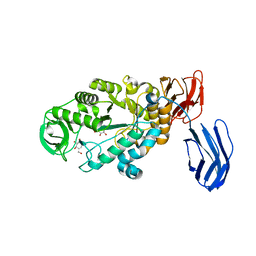 | | Alpha-amylase B in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3BCD
 
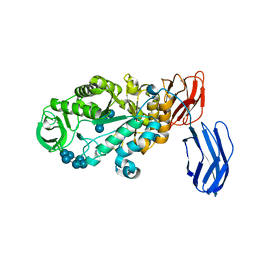 | | Alpha-amylase B in complex with maltotetraose and alpha-cyclodextrin | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3BCF
 
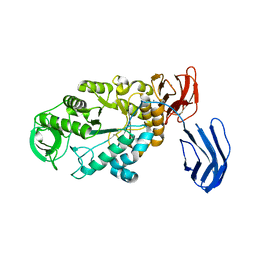 | | Alpha-amylase B from Halothermothrix orenii | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3FHV
 
 | |
5ZTZ
 
 | |
5ZU0
 
 | |
5ZZW
 
 | |
