2XN2
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
5MIP
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (30 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
2XN1
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
2XN0
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, PLATINUM (II) ION | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
5MIS
 
 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase Exposed to Visible Light (180 min) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
4C1U
 
 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with arabinoxylobiose | | Descriptor: | PENTAETHYLENE GLYCOL, SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fredslund, F, Ejby, M, Vujicic-Zagar, A, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
4BP3
 
 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with DECARBOXYLATED S-ADENOSYLMETHIONINE5' AND 4- METHYLANILINE | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-METHYLANILINE, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, ... | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2013-05-23 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4BP1
 
 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with 5'-Methylthioadenosine and Putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2013-05-23 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4CXM
 
 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with METHYLTHIOADENOSIN AND SPERMIDINE after catalysis in crystal | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, SPERMIDINE, SPERMIDINE SYNTHASE | | Authors: | Sprenger, J, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4CWA
 
 | | Structure of Plasmodium Falciparum Spermidine Synthase in Complex with 1H-Benzimidazole-2-pentanamine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5-(1H-benzimidazol-2-yl)pentan-1-amine, SPERMIDINE SYNTHASE | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4CVW
 
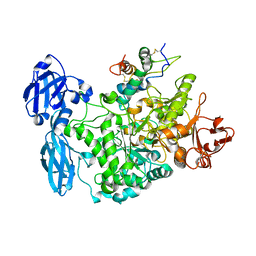 | | Structure of the barley limit dextrinase-limit dextrinase inhibitor complex | | Descriptor: | CALCIUM ION, LIMIT DEXTRINASE, LIMIT DEXTRINASE INHIBITOR | | Authors: | Moeller, M.S, Vester-Christensen, M.B, Jensen, J.M, Abou Hachem, M, Henriksen, A, Svensson, B. | | Deposit date: | 2014-03-31 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal Structure of Barley Limit Dextrinase:Limit Dextrinase Inhibitor (Ld:Ldi) Complex Reveals Insights Into Mechanism and Diversity of Cereal-Type Inhibitors.
J.Biol.Chem., 290, 2015
|
|
4C1T
 
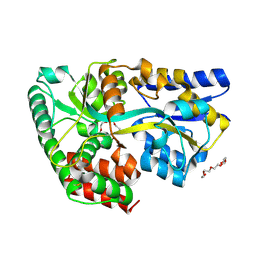 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with arabinoxylotriose | | Descriptor: | PENTAETHYLENE GLYCOL, SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, alpha-L-arabinofuranose-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fredslund, F, Ejby, M, Vujicic-Zagar, A, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
1GAH
 
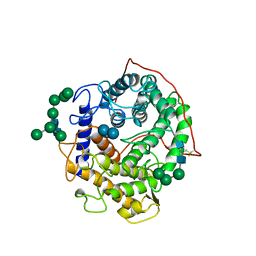 | | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
2VLT
 
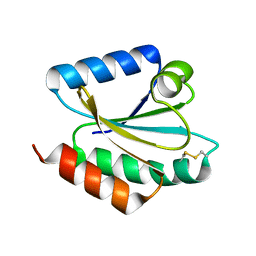 | | Crystal structure of barley thioredoxin h isoform 2 in the oxidized state | | Descriptor: | THIOREDOXIN H ISOFORM 2. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-16 | | Release date: | 2008-04-29 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2VLV
 
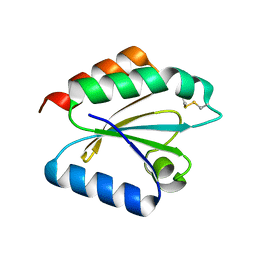 | | Crystal structure of barley thioredoxin h isoform 2 in partially radiation-reduced state | | Descriptor: | THIOREDOXIN H ISOFORM 2. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-16 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2VM1
 
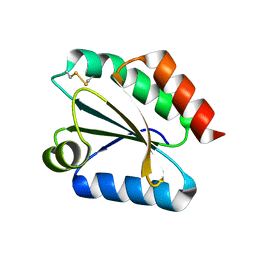 | | Crystal structure of barley thioredoxin h isoform 1 crystallized using ammonium sulfate as precipitant | | Descriptor: | SULFATE ION, THIOREDOXIN H ISOFORM 1. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-04-29 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2VLU
 
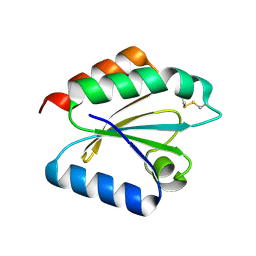 | | Crystal structure of barley thioredoxin h isoform 2 in partially radiation-reduced state | | Descriptor: | THIOREDOXIN H ISOFORM 2. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-16 | | Release date: | 2008-04-29 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2VM2
 
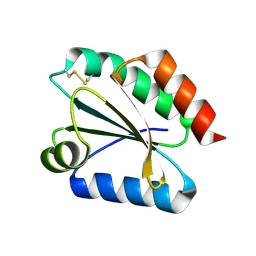 | | Crystal structure of barley thioredoxin h isoform 1 crystallized using PEG as precipitant | | Descriptor: | THIOREDOXIN H ISOFORM 1. | | Authors: | Maeda, K, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Barley Thioredoxin H Isoforms Hvtrxh1 and Hvtrxh2 Reveal Features Involved in Protein Recognition and Possibly in Discriminating the Isoform Specificity.
Protein Sci., 17, 2008
|
|
2WHD
 
 | | Barley NADPH-dependent thioredoxin reductase 2 | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE | | Authors: | Kirkensgaard, K.G, Hagglund, P, Finnie, C, Svensson, B, Henriksen, A. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Hordeum Vulgare Nadph-Dependent Thioredoxin Reductase 2. Unwinding the Reaction Mechanism.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
