1XOM
 
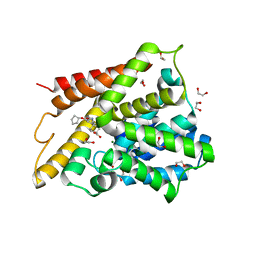 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Cilomilast | | Descriptor: | 1,2-ETHANEDIOL, CILOMILAST, MAGNESIUM ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
1Y2C
 
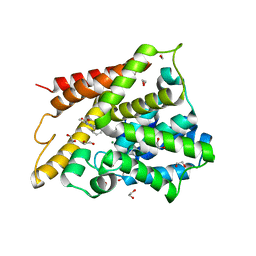 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 3,5-dimethyl-1-phenyl-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 1,2-ETHANEDIOL, 3,5-DIMETHYL-1-PHENYL-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
1Y2K
 
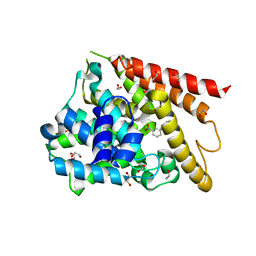 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 3,5-dimethyl-1-(3-nitro-phenyl)-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 1,2-ETHANEDIOL, 3,5-DIMETHYL-1-(3-NITROPHENYL)-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
2D3T
 
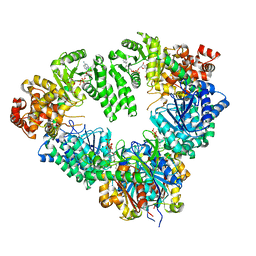 | | Fatty Acid beta-oxidation multienzyme complex from Pseudomonas Fragi, Form V | | Descriptor: | 3-ketoacyl-CoA thiolase, ACETYL COENZYME *A, Fatty oxidation complex alpha subunit, ... | | Authors: | Tsuchiya, D, Shimizu, N, Ishikawa, M, Suzuki, Y, Morikawa, K. | | Deposit date: | 2005-10-01 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-Induced Domain Rearrangement of Fatty Acid beta-Oxidation Multienzyme Complex
Structure, 14, 2006
|
|
2V0X
 
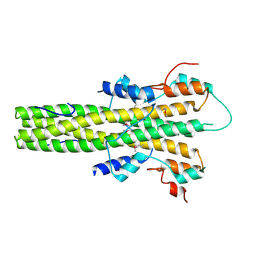 | | The dimerization domain of LAP2alpha | | Descriptor: | LAMINA-ASSOCIATED POLYPEPTIDE 2 ISOFORMS ALPHA/ZETA | | Authors: | Bradley, C.M, Jones, S, Huang, Y, Suzuki, Y, Kvaratskhelia, M, Hickman, A.B, Craigie, R, Dyda, F. | | Deposit date: | 2007-05-20 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Dimerization of Lap2Alpha, a Component of the Nuclear Lamina.
Structure, 15, 2007
|
|
4H53
 
 | | Influenza N2-Tyr406Asp neuraminidase in complex with beta-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
4H52
 
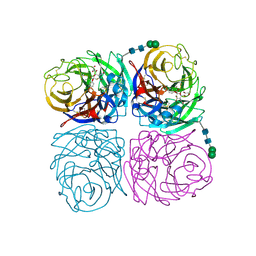 | | Wild-type influenza N2 neuraminidase covalent complex with 3-fluoro-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
6JTB
 
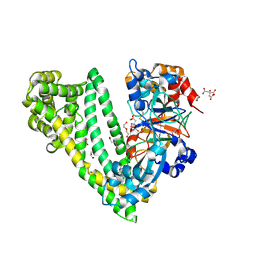 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with citrate from Porphyromonas gingivalis (Space) | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
6JTC
 
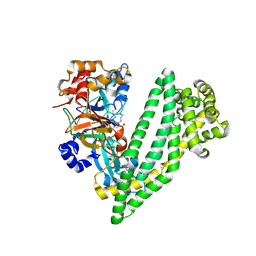 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with SH-5 from Porphyromonas gingivalis (Space) | | Descriptor: | 2-(2-azanylethylamino)-5-nitro-benzoic acid, Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
5YP4
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, LYSINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP1
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP2
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with DPP4 inhibitor from Pseudoxanthomonas mexicana WO24 | | Descriptor: | (2S,5R)-1-[2-[[1-(hydroxymethyl)cyclopentyl]amino]ethanoyl]pyrrolidine-2,5-dicarbonitrile, Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
5YP3
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, ISOLEUCINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
7DKD
 
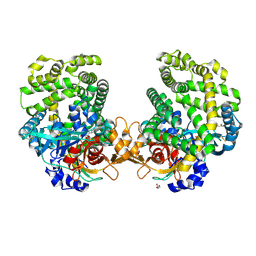 | | Stenotrophomonas maltophilia DPP7 in complex with Asn-Tyr | | Descriptor: | ASPARAGINE, Dipeptidyl-peptidase, GLYCEROL, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKC
 
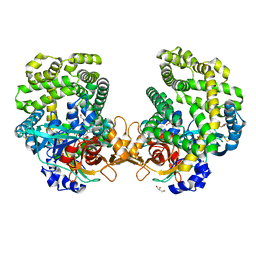 | | Stenotrophomonas maltophilia DPP7 in complex with Tyr-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, TYROSINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKE
 
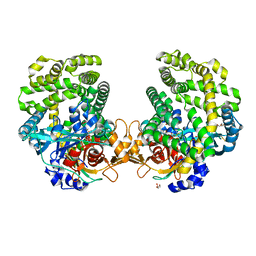 | | Stenotrophomonas maltophilia DPP7 in complex with Phe-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, PHENYLALANINE, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKB
 
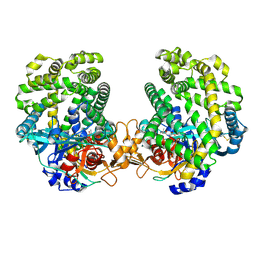 | | Stenotrophomonas maltophilia DPP7 in complex with Val-Tyr | | Descriptor: | Dipeptidyl-peptidase, TYROSINE, VALINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
5B2C
 
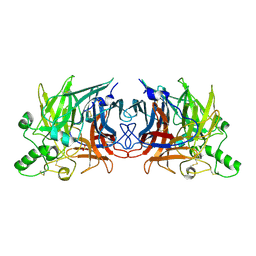 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, SULFATE ION | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B2D
 
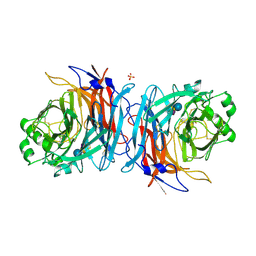 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to 3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1TB5
 
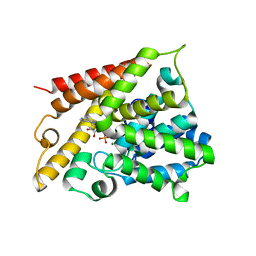 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
1TBF
 
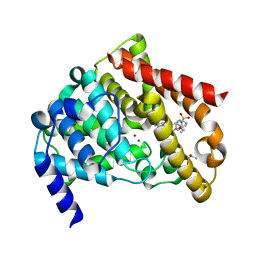 | | Catalytic Domain Of Human Phosphodiesterase 5A in Complex with Sildenafil | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
1T9R
 
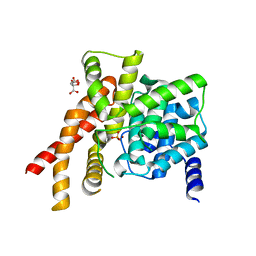 | | Catalytic Domain Of Human Phosphodiesterase 5A | | Descriptor: | CITRIC ACID, PHOSPHATE ION, ZINC ION, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-18 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
1TAZ
 
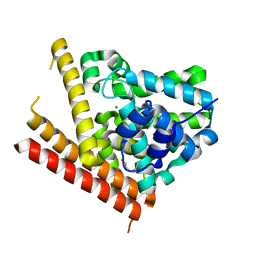 | | Catalytic Domain Of Human Phosphodiesterase 1B | | Descriptor: | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ZINC ION | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
3AH3
 
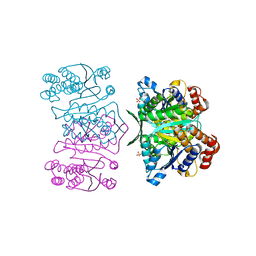 | | Crystal structure of LR5-1, 3-isopropylmalate dehydrogenase created by directed evolution | | Descriptor: | 1,2-ETHANEDIOL, Homoisocitrate dehydrogenase, SULFATE ION | | Authors: | Tomita, T, Suzuki, Y, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2010-04-13 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enhancement of the latent 3-isopropylmalate dehydrogenase activity of promiscuous homoisocitrate dehydrogenase by directed evolution
Biochem.J., 431, 2010
|
|
1T9S
 
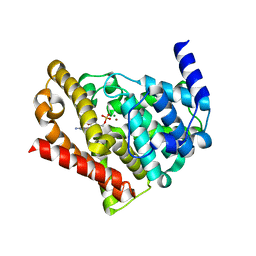 | | Catalytic Domain Of Human Phosphodiesterase 5A in Complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-18 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
