2CQM
 
 | | Solution structure of the mitochondrial ribosomal protein L17 isolog | | Descriptor: | Ribosomal protein L17 isolog | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mitochondrial ribosomal protein L17 isolog
To be Published
|
|
2CQI
 
 | | Solution structure of the RNA binding domain of Nucleolysin TIAR | | Descriptor: | Nucleolysin TIAR | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of Nucleolysin TIAR
To be Published
|
|
2CQH
 
 | | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2 | | Descriptor: | IGF-II mRNA-binding protein 2 isoform a | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2
To be Published
|
|
2CQJ
 
 | | Solution structure of the S4 domain of U3 small nucleolar ribonucleoprotein protein IMP3 homolog | | Descriptor: | U3 small nucleolar ribonucleoprotein protein IMP3 homolog | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the S4 domain of U3 small nucleolar ribonucleoprotein protein IMP3 homolog
To be Published
|
|
2CQK
 
 | | Solution structure of the La domain of c-Mpl binding protein | | Descriptor: | C-Mpl binding protein | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the La domain of c-Mpl binding protein
To be Published
|
|
2CQL
 
 | | Solution structure of the N-terminal domain of human ribosomal protein L9 | | Descriptor: | 60S ribosomal protein L9 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of human ribosomal protein L9
To be Published
|
|
2CQN
 
 | | Solution structure of the FF domain of human Formin-binding protein 3 | | Descriptor: | Formin-binding protein 3 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the FF domain of human Formin-binding protein 3
To be Published
|
|
2CQO
 
 | | Solution structure of the S1 RNA binding domain of human hypothetical protein FLJ11067 | | Descriptor: | Nucleolar protein of 40 kDa | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the S1 RNA binding domain of human hypothetical protein FLJ11067
To be Published
|
|
2CQP
 
 | | Solution structure of the RNA binding domain of RNA-binding protein 12 | | Descriptor: | RNA-binding protein 12 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of RNA-binding protein 12
To be Published
|
|
8IHI
 
 | | Cryo-EM structure of HCA2-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHH
 
 | | Cryo-EM structure of HCA2-Gi complex with LUF6283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-butyl-1~{H}-pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHF
 
 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
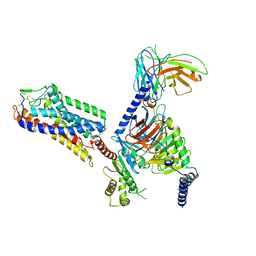 | | Cryo-EM structure of HCA3-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHB
 
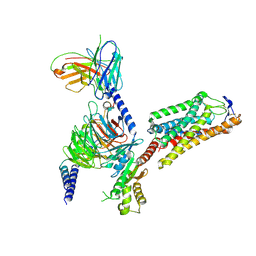 | | Cryo-EM structure of HCA2-Gi complex with GSK256073 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
2YQQ
 
 | | Solution structure of the zf-HIT domain in zinc finger HIT domain-containing protein 3 (TRIP-3) | | Descriptor: | ZINC ION, Zinc finger HIT domain-containing protein 3 | | Authors: | Suzuki, S, He, F, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-HIT domain in zinc finger HIT domain-containing protein 3 (TRIP-3)
To be Published
|
|
2ZW2
 
 | | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS) | | Descriptor: | GLYCEROL, Putative uncharacterized protein STS178 | | Authors: | Suzuki, S, Tamura, S, Okada, K, Baba, S, Kumasaka, T, Nakagawa, N, Masui, R, Kuramitsu, S, Sampei, G, Kawai, G. | | Deposit date: | 2008-11-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS)
To be Published
|
|
7Y13
 
 | | Cryo-EM structure of apo-state MrgD-Gi complex (local) | | Descriptor: | PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y12
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine | | Descriptor: | BETA-ALANINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y14
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine (local) | | Descriptor: | BETA-ALANINE, PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y15
 
 | | Cryo-EM structure of apo-state MrgD-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
3VIU
 
 | | Crystal structure of PurL from thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Suzuki, S, Yanai, H, Kanagawa, M, Tamura, S, Watanabe, Y, Fuse, K, Baba, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-10-12 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of N-formylglycinamide ribonucleotide amidotransferase II (PurL) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
3WNN
 
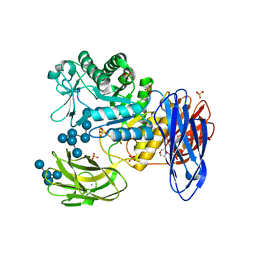 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
1UAT
 
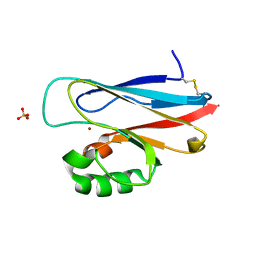 | | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J | | Descriptor: | Azurin iso-2, COPPER (II) ION, SULFATE ION | | Authors: | Inoue, T, Suzuki, S, Nisho, N, Yamaguchi, K, Kataoka, K, Tobari, J, Yong, X, Hamanaka, S, Matsumura, H, Kai, Y. | | Deposit date: | 2003-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J
J.Mol.Biol., 333, 2003
|
|
