6LS1
 
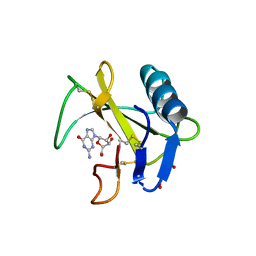 | | Ribonuclease from Hericium erinaceus active and GMP binding form | | 分子名称: | DI(HYDROXYETHYL)ETHER, GUANOSINE, Ribonuclease T1, ... | | 著者 | Takebe, K, Suzuki, M, Sangawa, T, Kobayashi, H, Itagaki, T. | | 登録日 | 2020-01-16 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Ribonuclease from Hericium erinaceus active and GMP binding form
To Be Published
|
|
5SW0
 
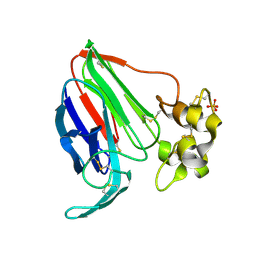 | |
5SW1
 
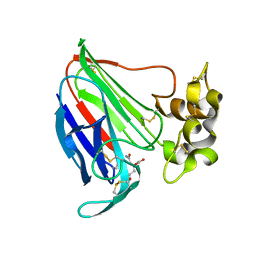 | | Thaumatin Structure at pH 6.0 | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Thaumatin I | | 著者 | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | 登録日 | 2016-08-08 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Thaumatin Structure at pH 6.0
To Be Published
|
|
5SW2
 
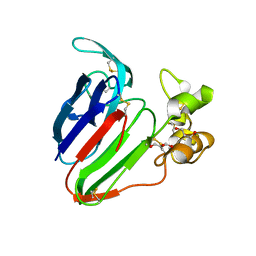 | | Thaumatin Structure at pH 6.0, orthorhombic type1 | | 分子名称: | GLYCEROL, Thaumatin I | | 著者 | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | 登録日 | 2016-08-08 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Thaumatin Structure at pH 6.0, orthorhombic type1
To Be Published
|
|
7XGI
 
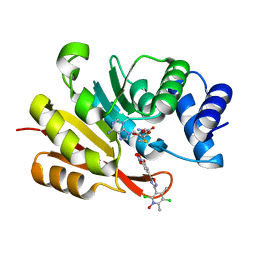 | | COMT SAH Mg opicapone complex | | 分子名称: | Catechol O-methyltransferase, MAGNESIUM ION, Opicapone, ... | | 著者 | Takebe, K, Kuwada-Kusunose, T, Suzuki, M, Iijima, H. | | 登録日 | 2022-04-04 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
7XJB
 
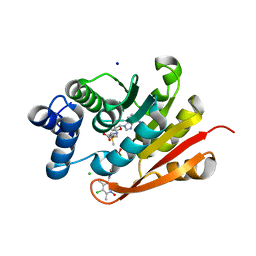 | | Rat-COMT, opicapone,SAM and Mg bond | | 分子名称: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | 著者 | Takebe, K, Iijima, H, Suzuki, M, Kuwada-Kusunose, T. | | 登録日 | 2022-04-15 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
1WR6
 
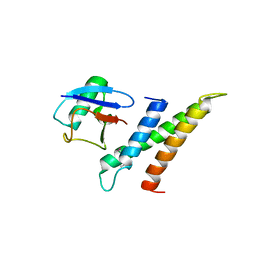 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | 分子名称: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | 著者 | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | 登録日 | 2004-10-12 | | 公開日 | 2005-06-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
1O3X
 
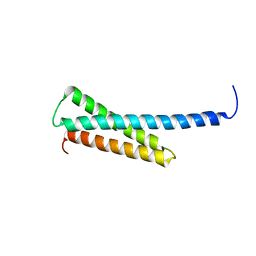 | | Crystal structure of human GGA1 GAT domain | | 分子名称: | ADP-ribosylation factor binding protein GGA1 | | 著者 | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | 登録日 | 2003-05-08 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular Mechanism of Membrane Recruitment of Gga by Arf in Lysosomal Protein Transport
Nat.Struct.Biol., 10, 2003
|
|
1O3Y
 
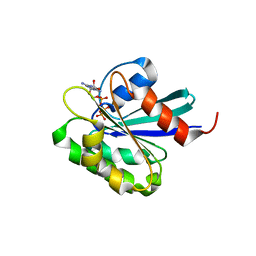 | | Crystal structure of mouse ARF1 (delta17-Q71L), GTP form | | 分子名称: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | 著者 | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | 登録日 | 2003-05-08 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
Nat.Struct.Biol., 10, 2003
|
|
2GCC
 
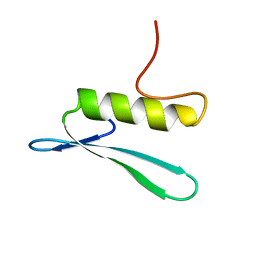 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | 分子名称: | ATERF1 | | 著者 | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | 登録日 | 1998-03-13 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
1GCC
 
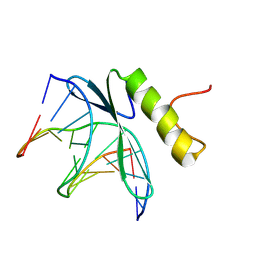 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF GCC-BOX BINDING DOMAIN OF ATERF1 AND GCC-BOX DNA, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*GP*CP*TP*GP*GP*CP*GP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*CP*GP*CP*CP*AP*GP*C)-3'), ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 | | 著者 | Yamasaki, K, Allen, M.D, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | 登録日 | 1998-03-13 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-10-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
8K6T
 
 | | The minor pilin structure of FctB3 in Streptococcus | | 分子名称: | FctB3, GLYCEROL | | 著者 | Takebe, K, Sangawa, T, Suzuki, M, Nakata, M. | | 登録日 | 2023-07-25 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Analysis of FctB3 crystal structure and insight into its structural stabilization and pilin linkage mechanisms.
Arch.Microbiol., 206, 2023
|
|
8JZ8
 
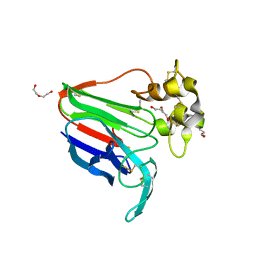 | | Subatomic structure of orthorhombic thaumatin at 0.89 Angstroms | | 分子名称: | DI(HYDROXYETHYL)ETHER, Thaumatin I | | 著者 | Masuda, T, Suzuki, M, Yamasaki, M, Mikami, B. | | 登録日 | 2023-07-04 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (0.89 Å) | | 主引用文献 | Subatomic structure of orthorhombic thaumatin at 0.89 angstrom reveals that highly flexible conformations are crucial for thaumatin sweetness.
Biochem.Biophys.Res.Commun., 703, 2024
|
|
1WZV
 
 | | Crystal Structure of UbcH8 | | 分子名称: | Ubiquitin-conjugating enzyme E2 L6 | | 著者 | Mizushima, T, Suzuki, M, Teshima, N, Yamane, T, Murata, S, Tanaka, K. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of UbcH8
To be Published
|
|
1WZW
 
 | | Crystal Structure of UbcH8 | | 分子名称: | Ubiquitin-conjugating enzyme E2 L6 | | 著者 | Mizushima, T, Suzuki, M, Teshima, N, Yamane, T, Murata, S, Tanaka, K. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of UbcH8
To be Published
|
|
1MP9
 
 | | TBP from a mesothermophilic archaeon, Sulfolobus acidocaldarius | | 分子名称: | TATA-binding protein | | 著者 | Koike, H, Kawashima-Ohya, Y, Yamasaki, T, Clowney, L, Katsuya, Y, Suzuki, M. | | 登録日 | 2002-09-12 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Origins of Protein Stability Revealed by Comparing Crystal Structures of TATA Binding Proteins.
Structure, 12, 2004
|
|
5D4H
 
 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | 分子名称: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-08-07 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4J
 
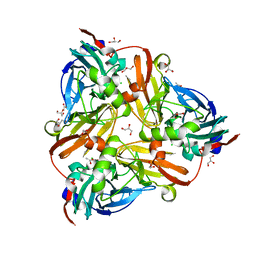 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | 分子名称: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-08-07 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4I
 
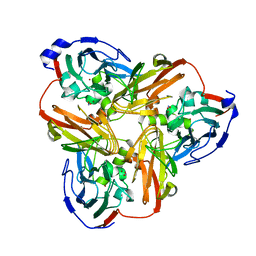 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | 著者 | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-08-07 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F7A
 
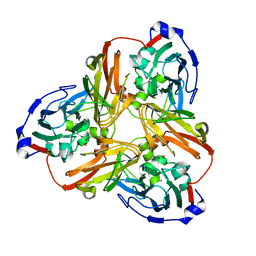 | | Nitrite complex structure of copper nitrite reductase from Alcaligenes faecalis determined at 293 K | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | 著者 | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-12-07 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F7B
 
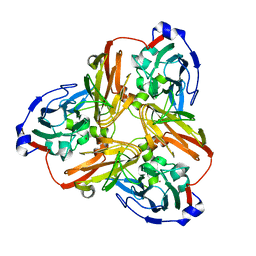 | | Resting state structure of CuNiR form Alcaligenes faecalis determined at 293 K | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase | | 著者 | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-12-07 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2F08
 
 | | Crystal structure of a major house dust mite allergen, Derf 2 | | 分子名称: | O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, mite allergen Der f II | | 著者 | Mikami, B, Tanaka, Y, Minato, N, Suzuki, M, Korematsu, S. | | 登録日 | 2005-11-12 | | 公開日 | 2005-11-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and some properties of a major house dust mite allergen, Derf 2
Biochem.Biophys.Res.Commun., 339, 2006
|
|
2HI7
 
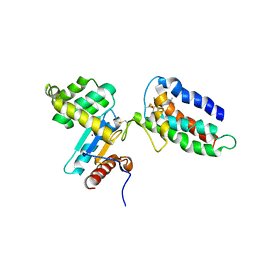 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | 分子名称: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | 著者 | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | 登録日 | 2006-06-29 | | 公開日 | 2006-12-05 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
2K7W
 
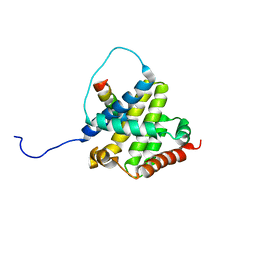 | | BAX Activation is Initiated at a Novel Interaction Site | | 分子名称: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | 著者 | Gavathiotis, E, Suzuki, M, Davis, M.L, Pitter, K, Bird, G.H, Katz, S.G, Tu, H.C, Kim, H, Cheng, E.H, Tjandra, N, Walensky, L.D. | | 登録日 | 2008-08-27 | | 公開日 | 2008-10-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | BAX activation is initiated at a novel interaction site.
Nature, 455, 2008
|
|
2E1A
 
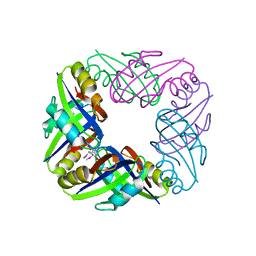 | | crystal structure of FFRP-DM1 | | 分子名称: | 75aa long hypothetical regulatory protein AsnC, SELENOMETHIONINE | | 著者 | Koike, H, Suzuki, M. | | 登録日 | 2006-10-19 | | 公開日 | 2007-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Structural Code for Discriminating between Transcription Signals Revealed by the Feast/Famine Regulatory Protein DM1 in Complex with Ligands
Structure, 15, 2007
|
|
