2AZJ
 
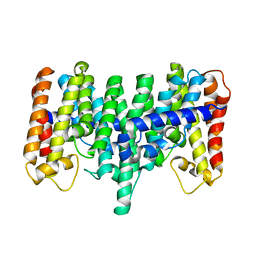 | | Crystal structure for the mutant D81C of Sulfolobus solfataricus hexaprenyl pyrophosphate synthase | | Descriptor: | Geranylgeranyl pyrophosphate synthetase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H.J. | | Deposit date: | 2005-09-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
4XPJ
 
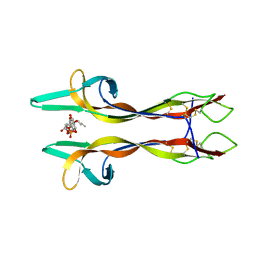 | | Crystal structure of Nerve growth factor in complex with lysophosphatidylinositol | | Descriptor: | (2R)-2-hydroxy-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl tridecanoate, Beta-nerve growth factor | | Authors: | Sun, H.L, Jiang, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | The structure of nerve growth factor in complex with lysophosphatidylinositol
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1MIF
 
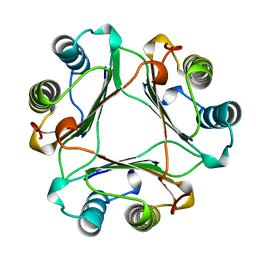 | | MACROPHAGE MIGRATION INHIBITORY FACTOR (MIF) | | Descriptor: | MACROPHAGE MIGRATION INHIBITORY FACTOR | | Authors: | Sun, H.-W, Lolis, E. | | Deposit date: | 1996-01-26 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
6M1V
 
 | |
5C71
 
 | | The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Glucuronidase, alpha-D-glucopyranuronic acid | | Authors: | Sun, H.L, Lv, B, Huang, S, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-guided engineering of the substrate specificity of a fungal beta-glucuronidase toward triterpenoid saponins.
J.Biol.Chem., 293, 2018
|
|
5WTD
 
 | | Structure of human serum transferrin bound ruthenium at N-lobe | | Descriptor: | FE (III) ION, MALONATE ION, RUTHENIUM ION, ... | | Authors: | Sun, H, Wang, M, Lai, T.P, Zhang, H, Hao, Q. | | Deposit date: | 2016-12-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
5C70
 
 | | The structure of Aspergillus oryzae beta-glucuronidase | | Descriptor: | Glucuronidase | | Authors: | Sun, H.L, Lv, B, Huang, S, Sun, Q.F, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Enhancing the Thermostability of beta-Glucuronidase by Rationally Redesigning the Catalytic Domain Based on Sequence Alignment Strategy
Ind Eng Chem Res, 55, 2016
|
|
7DEA
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck Northern China/22/2017 (H5N6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Wei, X, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
7DEB
 
 | | Structure of an avian influenza H5 hemagglutinin from the influenza virus A/duck/Eastern China/L0230/2010 (H5N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Sun, H, Sun, H, Song, J, Zhang, W, Qi, J, Gao, G.F, Liu, J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Haemagglutinin and neuraminidase acid stability in H5N6 avian influenza virus confers infection adaptation in mammals
To Be Published
|
|
5X5P
 
 | | Human serum transferrin bound to ruthenium NTA | | Descriptor: | FE (III) ION, MALONATE ION, NITRILOTRIACETIC ACID, ... | | Authors: | Sun, H, Wang, M. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
7XH0
 
 | | crystal structure of Csn-PD from Paenibacillus dendritiformis | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Chitosanase | | Authors: | Sun, H.H, Cheng, Y.M, Cao, R, Liu, Q, Zhao, L. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | crystal structure of Csn-PD from Paenibacillus dendritiformis
To Be Published
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | Descriptor: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | Authors: | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
8GTA
 
 | | Cryo-EM structure of the marine siphophage vB_Dshs-R4C capsid | | Descriptor: | Major capsid protein | | Authors: | Sun, H, Huang, Y, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
2DK9
 
 | | Solution structure of Calponin Homology domain of Human MICAL-1 | | Descriptor: | NEDD9-interacting protein with calponin homology and LIM domains | | Authors: | Sun, H, Dai, H, Zhang, J, Xiong, S, Wu, J, Shi, Y. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calponin homology domain of Human MICAL-1
J.Biomol.Nmr, 36, 2006
|
|
2AZK
 
 | | Crystal structure for the mutant W136E of Sulfolobus solfataricus hexaprenyl pyrophosphate synthase | | Descriptor: | Geranylgeranyl pyrophosphate synthetase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H.J. | | Deposit date: | 2005-09-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
2AZL
 
 | | Crystal structure for the mutant F117E of Thermotoga maritima octaprenyl pyrophosphate synthase | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H. | | Deposit date: | 2005-09-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
2NZY
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose | | Descriptor: | Alpha1,3-Fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Sun, H.Y, Ko, T.P. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design.
J. Biol. Chem., 282, 2007
|
|
2NZW
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase | | Descriptor: | Alpha1,3-fucosyltransferase, SULFATE ION | | Authors: | Sun, H.Y, Ko, T.P. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design.
J. Biol. Chem., 282, 2007
|
|
