3CXP
 
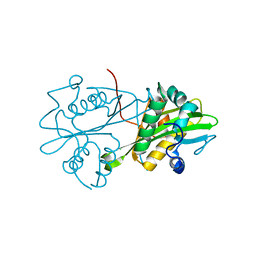 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 mutant E156A | | Descriptor: | CHLORIDE ION, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3SQZ
 
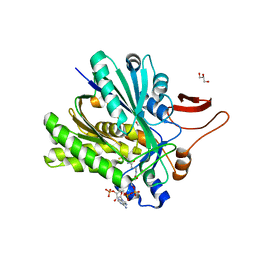 | |
6L55
 
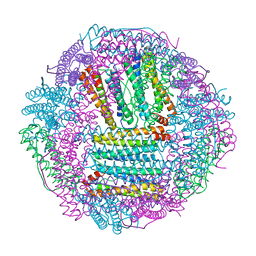 | | Recombinant Tegillarca granosa ferritin | | Descriptor: | FE (III) ION, FORMIC ACID, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78304863 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
3CXQ
 
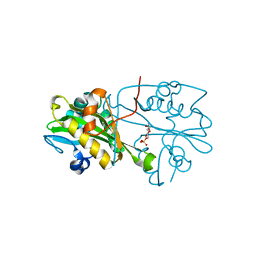 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3CXS
 
 | | Crystal structure of human GNA1 | | Descriptor: | Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
7VHR
 
 | | Apostichopus japonicus ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Wu, Y, Su, X.R, Ming, T.H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystallographic characterization of a marine invertebrate ferritin from the sea cucumber Apostichopus japonicus.
Febs Open Bio, 12, 2022
|
|
4F8Y
 
 | |
3B3D
 
 | | B.subtilis YtbE | | Descriptor: | CALCIUM ION, Putative morphine dehydrogenase | | Authors: | Zhou, Y.F, Li, L.F, Liang, Y.H, Su, X.-D. | | Deposit date: | 2007-10-20 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
3EXS
 
 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXT
 
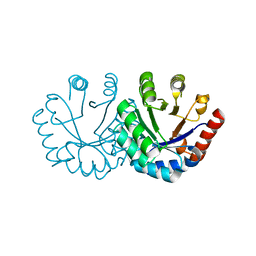 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | MAGNESIUM ION, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Liu, X, Li, G.L, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
2HVV
 
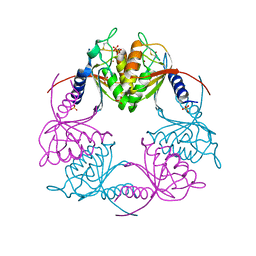 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | Descriptor: | SULFATE ION, ZINC ION, deoxycytidylate deaminase | | Authors: | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
2HVW
 
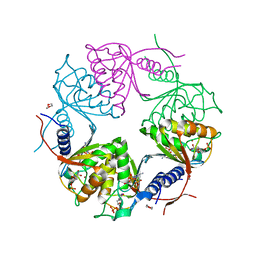 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 3,4-DIHYDRO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
2G0J
 
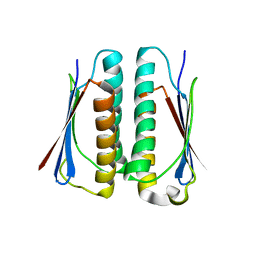 | | Crystal structure of SMU.848 from Streptococcus mutans | | Descriptor: | hypothetical protein SMU.848 | | Authors: | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3BR8
 
 | |
3H6X
 
 | |
3SR7
 
 | |
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
7QA9
 
 | | 10bp DNA/DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6L56
 
 | | Fe(II) loaded Tegillarca granosa ferritin | | Descriptor: | FE (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85300577 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
2G3F
 
 | | Crystal Structure of imidazolonepropionase complexed with imidazole-4-acetic acid sodium salt, a substrate homologue | | Descriptor: | 2H-IMIDAZOL-4-YLACETIC ACID, Imidazolonepropionase, ZINC ION | | Authors: | Yu, Y, Liang, Y.H, Su, X.D. | | Deposit date: | 2006-02-19 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A catalytic mechanism revealed by the crystal structures of the imidazolonepropionase from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
3V6L
 
 | | Crystal Structure of caspase-6 inactivation mutation | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
3V6M
 
 | | Inhibition of caspase-6 activity by single mutation outside the active site | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
3E4O
 
 | | Crystal structure of succinate bound state DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MAGNESIUM ION, SUCCINIC ACID | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
7EK0
 
 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | Descriptor: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
