3RU8
 
 | |
3SAO
 
 | | The Siderocalin Ex-FABP functions through dual ligand specificities | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2,3-DIHYDROXY-BENZOIC ACID, Extracellular fatty acid-binding protein, ... | | Authors: | Correnti, C, Strong, R.K, Clifton, M.C. | | Deposit date: | 2011-06-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Galline Ex-FABP Is an Antibacterial Siderocalin and a Lysophosphatidic Acid Sensor Functioning through Dual Ligand Specificities.
Structure, 19, 2011
|
|
3S26
 
 | | Crystal Structure of Murine Siderocalin (Lipocalin 2, 24p3) | | Descriptor: | Neutrophil gelatinase-associated lipocalin, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Correnti, C, Bandaranayake, A.D, Strong, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-28 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors.
Nucleic Acids Res., 39, 2011
|
|
3T43
 
 | |
3U9P
 
 | | Crystal Structure of Murine Siderocalin in Complex with an Fab Fragment | | Descriptor: | Monoclonal Fab Fragment Heavy Chain, Monoclonal Fab Fragment Light Chain, Neutrophil gelatinase-associated lipocalin | | Authors: | Correnti, C, Strong, R.K. | | Deposit date: | 2011-10-19 | | Release date: | 2013-05-01 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Siderocalin/Lcn2/NGAL/24p3 does not drive apoptosis through gentisic acid mediated iron withdrawal in hematopoietic cell lines.
Plos One, 7, 2012
|
|
1QQS
 
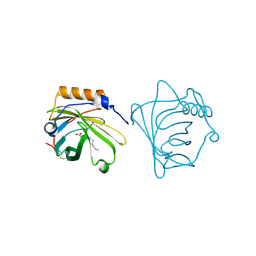 | | NEUTROPHIL GELATINASE ASSOCIATED LIPOCALIN HOMODIMER | | Descriptor: | DECANOIC ACID, NEUTROPHIL GELATINASE, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Goetz, D.H, Willie, S.T, Armen, R, Bratt, T, Borregaard, N, Strong, R.K. | | Deposit date: | 1999-06-07 | | Release date: | 2000-04-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand preference inferred from the structure of neutrophil gelatinase associated lipocalin
Biochemistry, 39, 2000
|
|
1L6M
 
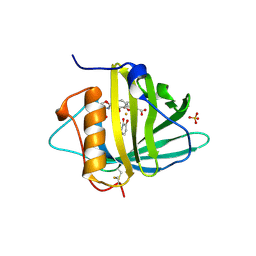 | | Neutrophil Gelatinase-associated Lipocalin is a Novel Bacteriostatic Agent that Interferes with Siderophore-mediated Iron Acquisition | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, FE (III) ION, ... | | Authors: | Goetz, D.H, Borregaard, N, Bluhm, M.E, Raymond, K.N, Strong, R.K. | | Deposit date: | 2002-03-11 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Neutrophil Lipocalin NGAL is a Bacteriostatic Agent that Interferes with Siderophore-mediated Iron Acquisition
Mol.Cell, 10, 2002
|
|
3FW5
 
 | |
3FW4
 
 | |
4JLR
 
 | | Crystal structure of a designed Respiratory Syncytial Virus Immunogen in complex with Motavizumab | | Descriptor: | Motavizumab Fab heavy chain, Motavizumab Fab light chain, PENTAETHYLENE GLYCOL, ... | | Authors: | Rupert, P.B, Correia, B, Schief, W, Strong, R.K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-02-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Proof of principle for epitope-focused vaccine design.
Nature, 507, 2014
|
|
4K19
 
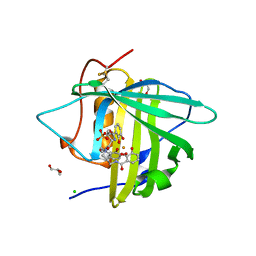 | | The structure of Human Siderocalin bound to the bacterial siderophore fluvibactin | | Descriptor: | (4S,5R)-N,N-bis{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, CHLORIDE ION, FE (III) ION, ... | | Authors: | Correnti, C, Clifton, M.C, Strong, R.K. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Siderocalin Outwits the Coordination Chemistry of Vibriobactin, a Siderophore of Vibrio cholerae.
Acs Chem.Biol., 8, 2013
|
|
6AVC
 
 | |
6AV8
 
 | |
6AU7
 
 | | Exploring Cystine Dense Peptide Space to Open a Unique Molecular Toolbox | | Descriptor: | GLYCEROL, Potassium channel toxin gamma-KTx 2.2, SULFATE ION | | Authors: | Gewe, M.M, Rupert, P, Strong, R.K. | | Deposit date: | 2017-08-30 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Screening, large-scale production and structure-based classification of cystine-dense peptides.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6AVA
 
 | |
6ATW
 
 | |
1X8U
 
 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with Carboxymycobactin T | | Descriptor: | CARBOXYMYCOBACTIN T, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-18 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
1X71
 
 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with TRENCAM-3,2-HOPO, a cepabactin analogue | | Descriptor: | 2,3-DIHYDROXYBENZAMIDE, FE (III) ION, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-12 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
1X89
 
 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with Carboxymycobactin S | | Descriptor: | CARBOXYMYCOBACTIN S, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
6ATN
 
 | |
6AVD
 
 | |
6ATS
 
 | |
6ATU
 
 | |
6AUP
 
 | | Exploring Cystine Dense Peptide Space to Open a Unique Molecular Toolbox | | Descriptor: | GLYCEROL, Potassium channel toxin gamma-KTx 2.2, SULFATE ION | | Authors: | Gewe, M.M, Rupert, P, Strong, R.K. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Screening, large-scale production and structure-based classification of cystine-dense peptides.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6ATL
 
 | | Exploring Cystine Dense Peptide Space to Open a Unique Molecular Toolbox | | Descriptor: | CITRIC ACID, Potassium channel toxin alpha-KTx 4.2, SULFATE ION | | Authors: | Gewe, M.M, Rupert, P, Strong, R.K. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Screening, large-scale production and structure-based classification of cystine-dense peptides.
Nat. Struct. Mol. Biol., 25, 2018
|
|
