1FEB
 
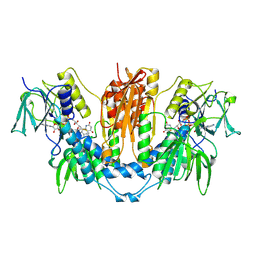 | |
1FEC
 
 | |
1FEA
 
 | |
9DOM
 
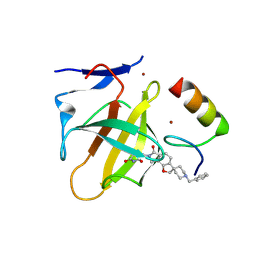 | | PVTX-405: A Potent, Highly Selective, and Orally Efficacious Molecular Glue Degrader of IKZF2 for Cancer Immunotherapy | | Descriptor: | (3S)-3-(1'-benzyl-6-oxo-6,8-dihydro-2H,7H-spiro[furo[2,3-e]isoindole-3,4'-piperidin]-7-yl)piperidine-2,6-dione, Protein cereblon, ZINC ION, ... | | Authors: | Strickland, C.O, Rice, C.T. | | Deposit date: | 2024-09-19 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of PVTX-405 as a potent and highly selective molecular glue degrader of IKZF2 for cancer immunotherapy.
Nat Commun, 16, 2025
|
|
2BED
 
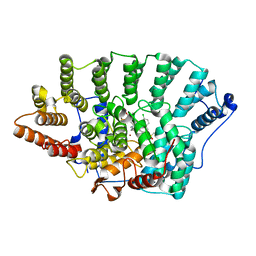 | | Structure of FPT bound to inhibitor SCH207736 | | Descriptor: | (11S)-8-CHLORO-11-[1-(METHYLSULFONYL)PIPERIDIN-4-YL]-6-PIPERAZIN-1-YL-11H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDINE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase beta subunit, ... | | Authors: | Strickland, C. | | Deposit date: | 2005-10-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced FTase activity achieved via piperazine interaction with catalytic zinc.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
9E1K
 
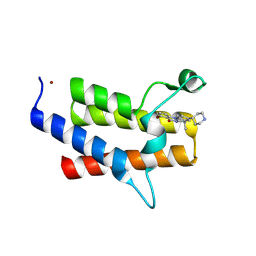 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 11 | | Descriptor: | (12S)-4-bromo-7,7-dimethyl-9-(piperidin-4-yl)indolo[1,2-a]quinazolin-5(7H)-one, Isoform Short of Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
9E31
 
 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 6 | | Descriptor: | (12'R)-4'-chloro-9'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, ACETATE ION, Isoform Short of Probable global transcription activator SNF2L2, ... | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-23 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
9E30
 
 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 13 | | Descriptor: | (12'S)-4'-chloro-10'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, ACETATE ION, Isoform Short of Probable global transcription activator SNF2L2, ... | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-23 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
2QMD
 
 | | Structure of BACE Bound to SCH722924 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4R)-4-(BENZYLOXY)PYRROLIDIN-2-YL]-1-(3,5-DIFLUOROBENZYL)-2-HYDROXYETHYL]-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C.O, Iserloh, U. | | Deposit date: | 2007-07-16 | | Release date: | 2008-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Potent pyrrolidine- and piperidine-based BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2QMG
 
 | | Structure of BACE Bound to SCH745966 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-2-[(2R,4R)-4-PHENOXYPYRROLIDIN-2-YL]ETHYL}-3-{[(2R)-2-(METHOXYMETHYL)PYRROLIDIN-1-YL]CARBONYL}-5-METHYLBENZAMIDE | | Authors: | Strickland, C.O, Iserloh, U. | | Deposit date: | 2007-07-16 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of an orally efficaceous 4-phenoxypyrrolidine-based BACE-1 inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3LPJ
 
 | | Structure of BACE Bound to SCH743641 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-[(1S,2S)-2-[(2R)-4-benzylpiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2R2L
 
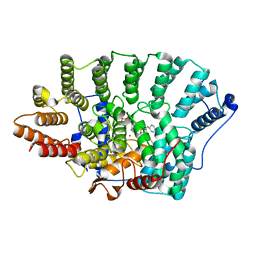 | | Structure of Farnesyl Protein Transferase bound to PB-93 | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyltransferase subunit alpha, Farnesyltransferase subunit beta, ... | | Authors: | Strickland, C.O, Voorhis, W. | | Deposit date: | 2007-08-27 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Efficacy, pharmacokinetics, and metabolism of tetrahydroquinoline inhibitors of Plasmodium falciparum protein farnesyltransferase.
Antimicrob.Agents Chemother., 51, 2007
|
|
3KN0
 
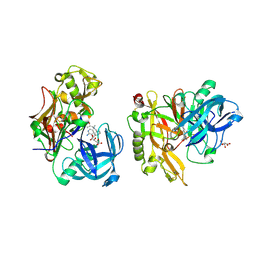 | | Structure of BACE bound to SCH708236 | | Descriptor: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3L5D
 
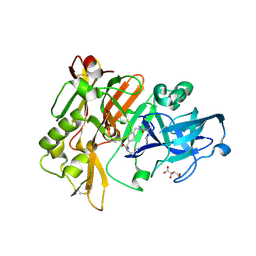 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5C
 
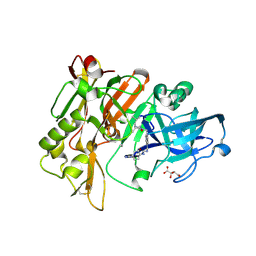 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L58
 
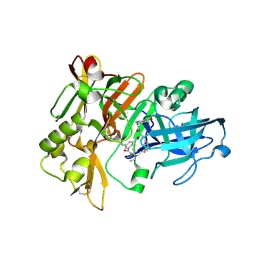 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3LPI
 
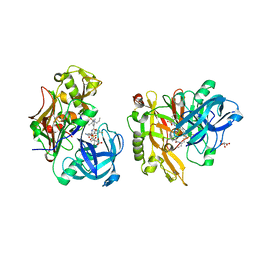 | | Structure of BACE Bound to SCH745132 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPK
 
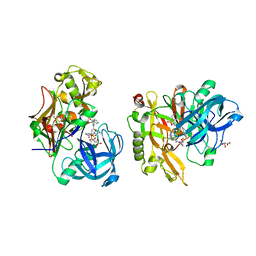 | | Structure of BACE Bound to SCH747123 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L5F
 
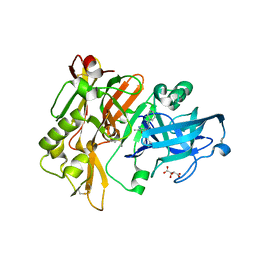 | | Structure of BACE Bound to SCH736201 | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-(cyclohexylmethyl)-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3CIB
 
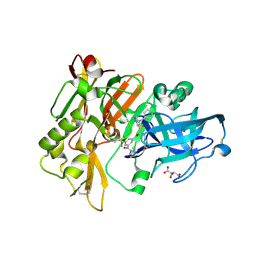 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CID
 
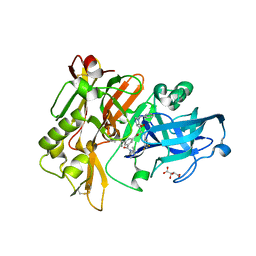 | | Structure of BACE Bound to SCH726222 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(4S)-1-benzyl-5-oxoimidazolidin-4-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CIC
 
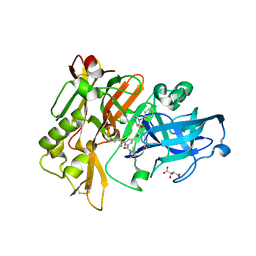 | | Structure of BACE Bound to SCH709583 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4DJW
 
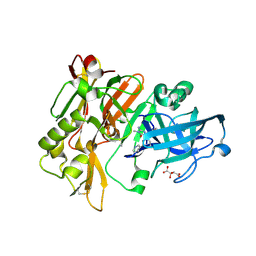 | |
4DJU
 
 | | Structure of BACE Bound to 2-imino-3-methyl-5,5-diphenylimidazolidin-4-one | | Descriptor: | (2E)-2-imino-3-methyl-5,5-diphenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DJV
 
 | | Structure of BACE Bound to 2-imino-5-(3'-methoxy-[1,1'-biphenyl]-3-yl)-3-methyl-5-phenylimidazolidin-4-one | | Descriptor: | (2E,5R)-2-imino-5-(3'-methoxybiphenyl-3-yl)-3-methyl-5-phenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
