6S30
 
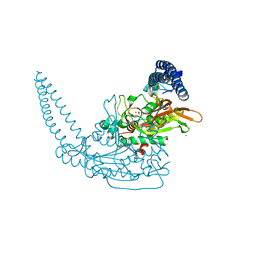 | | Crystal Structure of Seryl-tRNA Synthetase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Serine--tRNA ligase | | Authors: | Pang, L, De Graef, S, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
2XZ5
 
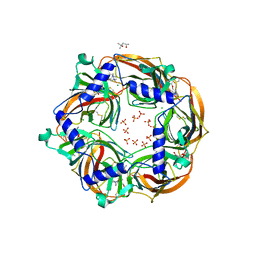 | | MMTS-modified Y53C mutant of Aplysia AChBP in complex with acetylcholine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE, ... | | Authors: | Brams, M, Gay, E.A, Colon Saez, J, Guskov, A, van Elk, R, van der Schors, R.C, Peigneur, S, Tytgat, J, Strelkov, S.V, Smit, A.B, Yakel, J.L, Ulens, C. | | Deposit date: | 2010-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of a Cysteine-Modified Mutant in Loop D of Acetylcholine Binding Protein
J.Biol.Chem., 286, 2011
|
|
6TTJ
 
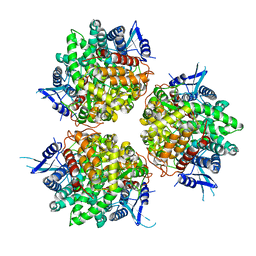 | | Neutral invertase 2 from Arabidopsis thaliana | | Descriptor: | Alkaline/neutral invertase CINV1 | | Authors: | Tsirkone, V.G, Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2019-12-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | Crystal structure of Arabidopsis thaliana neutral invertase 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2XZ6
 
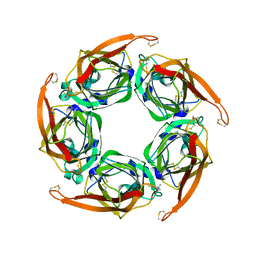 | | MTSET-modified Y53C mutant of Aplysia AChBP | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Gay, E.A, Colon Saez, J, Guskov, A, Van Elk, R, Van Der Schors, R.C, Peigneur, S, Tytgat, J, Strelkov, S.V, Smit, A.B, Yakel, J.L, Ulens, C. | | Deposit date: | 2010-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.137 Å) | | Cite: | Crystal Structures of a Cysteine-Modified Mutant in Loop D of Acetylcholine Binding Protein
J.Biol.Chem., 286, 2011
|
|
4DY3
 
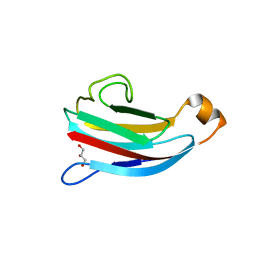 | | crystal structure of Escherichia coli PliG, a periplasmic lysozyme inhibitor of g-type lysozyme | | Descriptor: | GLYCEROL, Inhibitor of g-type lysozyme | | Authors: | Leysen, S, Vanheuverzwijn, S, Van Asten, K, Vanderkelen, L, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the PliG lysozyme inhibitor family.
J.Struct.Biol., 180, 2012
|
|
4G2U
 
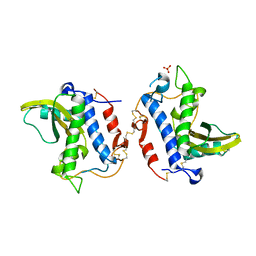 | | Crystal Structure Analysis of Ostertagia ostertagi ASP-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ancylostoma-secreted protein-like protein, SULFATE ION | | Authors: | Weeks, S.D, Borloo, J, Geldhof, P, Vercruysse, J, Strelkov, S.V. | | Deposit date: | 2012-07-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ostertagia ostertagi ASP-1: insights into disulfide-mediated cyclization and dimerization
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2Z79
 
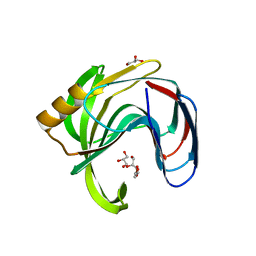 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | Descriptor: | Endo-1,4-beta-xylanase A, GLYCEROL | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2Y2W
 
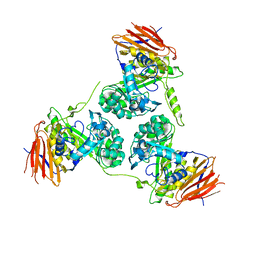 | | Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. | | Descriptor: | ARABINOFURANOSIDASE | | Authors: | Lagaert, S, Schoepe, J, Delcour, J.A, Lavigne, R, Strelkov, S.V, Courtin, C.M, Mikkelsen, N.E, Sandgren, M, Volckaert, G. | | Deposit date: | 2010-12-16 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elucidation of the Substrate Specificity and Protein Structure of Abfb, a Family 51 Alpha-L- Arabinofuranosidase from Bifidobacterium Longum.
To be Published
|
|
3Q9P
 
 | | HspB1 fragment | | Descriptor: | Heat shock protein beta-1 | | Authors: | Baranova, E.V, Beelen, S, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2011-01-09 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-Dimensional Structure of alpha-Crystallin Domain Dimers of Human Small Heat Shock Proteins HSPB1 and HSPB6
J.Mol.Biol., 2011
|
|
3Q9Q
 
 | |
4G9S
 
 | | Crystal structure of Escherichia coli PliG in complex with Atlantic salmon g-type lysozyme | | Descriptor: | CHLORIDE ION, CITRATE ANION, Goose-type lysozyme, ... | | Authors: | Leysen, S, Vanderkelen, L, Weeks, S.D, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-07-24 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis of bacterial defense against g-type lysozyme-based innate immunity.
Cell.Mol.Life Sci., 70, 2013
|
|
4DXZ
 
 | | crystal structure of a PliG-Ec mutant, a periplasmic lysozyme inhibitor of g-type lysozyme from Escherichia coli | | Descriptor: | GLYCEROL, Inhibitor of g-type lysozyme, SODIUM ION | | Authors: | Leysen, S, Vanheuverzwijn, S, Van Asten, K, Vanderkelen, L, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | crystal structure of a PliG-Ec mutant, a periplasmic lysozyme inhibitor of g-type lysozyme from Escherichia coli
To be Published
|
|
4DY5
 
 | | Crystal structure of Salmonella Typhimurium PliG, a periplasmic lysozyme inhibitor of g-type lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Gifsy-1 prophage protein | | Authors: | Leysen, S, Theuwis, V, Vanderkelen, L, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural characterization of the PliG lysozyme inhibitor family.
J.Struct.Biol., 180, 2012
|
|
1USD
 
 | | human VASP tetramerisation domain L352M | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Kuhnel, K, Jarchau, T, Wolf, E, Schlichting, I, Walter, U, Wittinghofer, A, Strelkov, S.V. | | Deposit date: | 2003-11-21 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Vasp Tetramerization Domain is a Right-Handed Coiled Coil Based on a 15-Residue Repeat
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3OE3
 
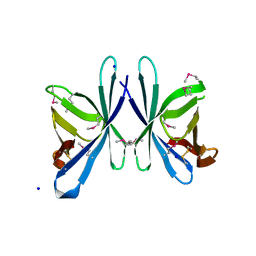 | | Crystal structure of PliC-St, periplasmic lysozyme inhibitor of C-type lysozyme from Salmonella typhimurium | | Descriptor: | Putative periplasmic protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-12 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
3OD9
 
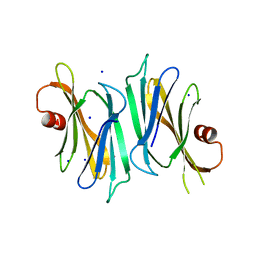 | | Crystal structure of PliI-Ah, periplasmic lysozyme inhibitor of I-type lysozyme from Aeromonas hydrophyla | | Descriptor: | POTASSIUM ION, Putative exported protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-11 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
3C7O
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with cellotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3S4R
 
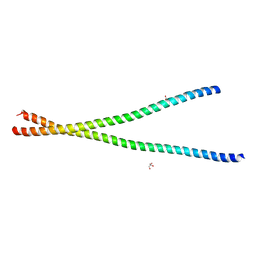 | |
3C7H
 
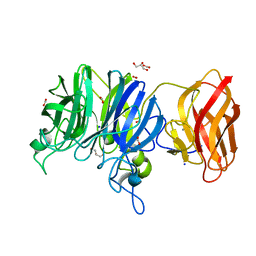 | | Crystal structure of glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with AXOS-4-0.5. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7E
 
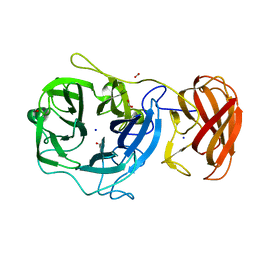 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7G
 
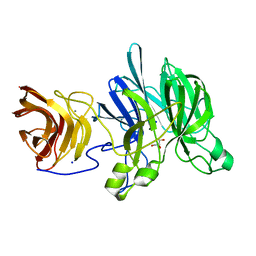 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with xylotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3UGF
 
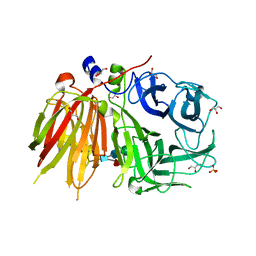 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3C7F
 
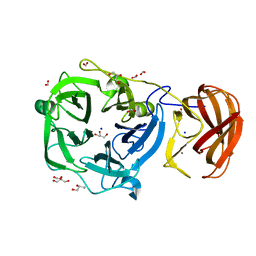 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
7AP4
 
 | | Thermus thermophilus Aspartyl-tRNA Synthetase in Complex with Compound AspS7HMDDA | | Descriptor: | (3~{S})-3-azanyl-4-[[(2~{R},3~{S},4~{R},5~{R})-5-[7-azanyl-5-(hydroxymethyl)benzimidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxysulfonylamino]-4-oxidanylidene-butanoic acid, Aspartate--tRNA(Asp) ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
6SNZ
 
 | | Crystal structure of lamin A coil1b tetramer | | Descriptor: | GLYCEROL, Prelamin-A/C | | Authors: | Lilina, A.V, Chernyatina, A.A, Guzenko, D, Strelkov, S.V. | | Deposit date: | 2019-08-28 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lateral A11type tetramerization in lamins.
J.Struct.Biol., 209, 2020
|
|
