1ED9
 
 | | STRUCTURE OF E. COLI ALKALINE PHOSPHATASE WITHOUT THE INORGANIC PHOSPHATE AT 1.75A RESOLUTION | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Stec, B, Holtz, K.M, Kantrowitz, E.R. | | Deposit date: | 2000-01-27 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A revised mechanism for the alkaline phosphatase reaction involving three metal ions.
J.Mol.Biol., 299, 2000
|
|
6NR7
 
 | | Rerefinement of chicken vinculin | | Descriptor: | (1R,2R,3S,4R,5R,6S)-4-{[(S)-[(2S)-2,3-dihydroxypropoxy](hydroxy)phosphoryl]oxy}-3,5,6-trihydroxycyclohexane-1,2-diyl bis[dihydrogen (phosphate)], PHOSPHATE ION, SULFATE ION, ... | | Authors: | Stec, B. | | Deposit date: | 2019-01-23 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refined model of chicken vinculin suggests the mechanism of activation by helical super-bundle unfurling
To be published
|
|
6NAK
 
 | | BACTERIAL PROTEIN COMPLEX TM BDE complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, TsaE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-05 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
1PHN
 
 | |
1ZG6
 
 | | TEM1 beta lactamase mutant S70G | | Descriptor: | Beta-lactamase TEM | | Authors: | Stec, B, Holtz, K.M, Wojciechowski, C.L, Kantrowitz, E.R. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the wild-type TEM-1 beta-lactamase at 1.55 A and the mutant enzyme Ser70Ala at 2.1 A suggest the mode of noncovalent catalysis for the mutant enzyme.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZG4
 
 | | TEM1 beta lactamase | | Descriptor: | Beta-lactamase TEM | | Authors: | Stec, B, Holtz, K.M, Wojciechowski, C.L, Kantrowitz, E.R. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the wild-type TEM-1 beta-lactamase at 1.55 A and the mutant enzyme Ser70Ala at 2.1 A suggest the mode of noncovalent catalysis for the mutant enzyme.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6NBJ
 
 | | Qri7 | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-07 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
2QG9
 
 | | Structure of a regulatory subunit mutant D19A of ATCase from E. coli | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Stec, B, Williams, M.K, Stieglitz, K.A, Kantrowitz, E.R. | | Deposit date: | 2007-06-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of two T-state structures of regulatory-chain mutants of Escherichia coli aspartate transcarbamoylase suggests that His20 and Asp19 modulate the response to heterotropic effectors.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QGF
 
 | | Structure of regulatory chain mutant H20A of asparate transcarbamoylase from E. coli | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Stec, B, Williams, M.K, Stieglitz, K.A, Kantrowitz, E.R. | | Deposit date: | 2007-06-28 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of two T-state structures of regulatory-chain mutants of Escherichia coli aspartate transcarbamoylase suggests that His20 and Asp19 modulate the response to heterotropic effectors.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1DK4
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
1ED8
 
 | | STRUCTURE OF E. COLI ALKALINE PHOSPHATASE INHIBITED BY THE INORGANIC PHOSPHATE AT 1.75A RESOLUTION | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Holtz, K.M, Kantrowitz, E.R. | | Deposit date: | 2000-01-27 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A revised mechanism for the alkaline phosphatase reaction involving three metal ions.
J.Mol.Biol., 299, 2000
|
|
1ELZ
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102G) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
1ELY
 
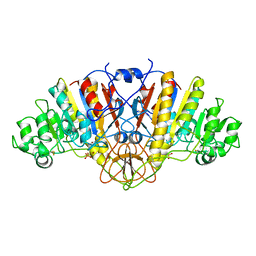 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102C) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
1ELX
 
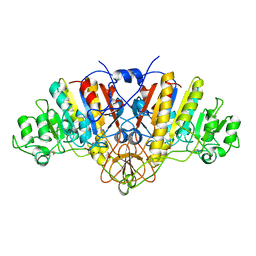 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102A) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
3MK0
 
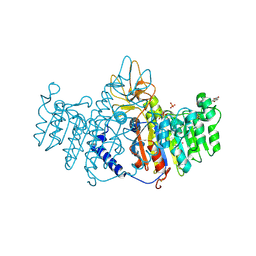 | | Refinement of placental alkaline phosphatase complexed with nitrophenyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alkaline phosphatase, ... | | Authors: | Stec, B, Cheltsov, A, Millan, J.L. | | Deposit date: | 2010-04-13 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3MK2
 
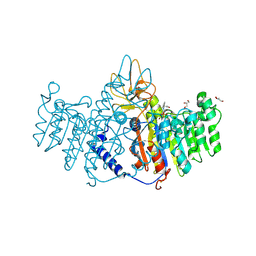 | | Placental alkaline phosphatase complexed with Phe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alkaline phosphatase, ... | | Authors: | Stec, B, Cheltsov, A, Millan, J.L. | | Deposit date: | 2010-04-13 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3MK1
 
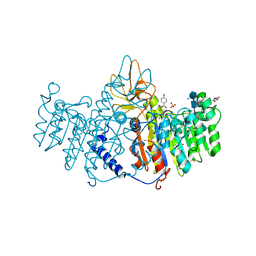 | | Refinement of placental alkaline phosphatase complexed with nitrophenyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alkaline phosphatase, ... | | Authors: | Stec, B, Cheltsov, A, Millan, J.L. | | Deposit date: | 2010-04-13 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1RDX
 
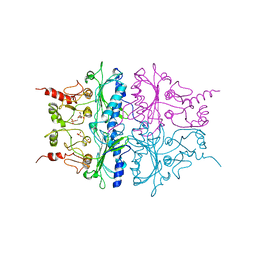 | | R-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1RDY
 
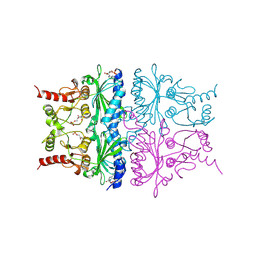 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
1RDZ
 
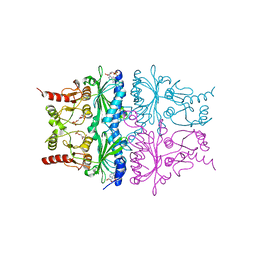 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
4F0H
 
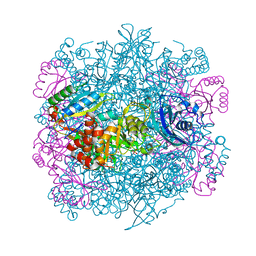 | | UNACTIVATED RUBISCO with OXYGEN BOUND | | Descriptor: | OXYGEN MOLECULE, PHOSPHATE ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Stec, B. | | Deposit date: | 2012-05-04 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanism of RuBisCO activation by carbamylation of the active site lysine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F0M
 
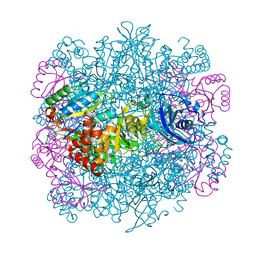 | | UNACTIVATED RUBISCO with MAGNESIUM AND A WATER MOLECULE BOUND | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Stec, B. | | Deposit date: | 2012-05-04 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism of RuBisCO activation by carbamylation of the active site lysine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F0K
 
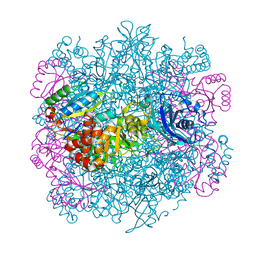 | | UNACTIVATED RUBISCO with MAGNESIUM AND CARBON DIOXIDE BOUND | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stec, B. | | Deposit date: | 2012-05-04 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism of RuBisCO activation by carbamylation of the active site lysine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F8B
 
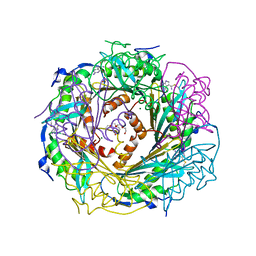 | | Crystal Structure of the Covalent Thioimide Intermediate of Unimodular Nitrile Reductase QueF | | Descriptor: | 2-amino-5-[(Z)-iminomethyl]-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MAGNESIUM ION, ... | | Authors: | Stec, B, Swairjo, M.A. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis of biological nitrile reduction.
J.Biol.Chem., 287, 2012
|
|
4FGC
 
 | | Crystal Structure of Active Site Mutant C55A of Nitrile Reductase QueF, Bound to Substrate PreQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, ... | | Authors: | Stec, B, Swairjo, M.A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structural basis of biological nitrile reduction.
J.Biol.Chem., 287, 2012
|
|
