2J0W
 
 | | Crystal structure of E. coli aspartokinase III in complex with aspartate and ADP (R-state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Kotaka, M, Ren, J, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-07 | | Release date: | 2006-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of R- and T-State Escherichia Coli Aspartokinase III: Mechanisms of the Allosteric Transition and Inhibition by Lysine.
J.Biol.Chem., 281, 2006
|
|
2GEM
 
 | |
2J0X
 
 | | CRYSTAL STRUCTURE OF E. COLI ASPARTOKINASE III IN COMPLEX WITH LYSINE AND ASPARTATE (T-STATE) | | Descriptor: | ASPARTIC ACID, LYSINE, LYSINE-SENSITIVE ASPARTOKINASE 3, ... | | Authors: | Kotaka, M, Ren, J, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-07 | | Release date: | 2006-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of R- and T-State Escherichia Coli Aspartokinase III: Mechanisms of the Allosteric Transition and Inhibition by Lysine.
J.Biol.Chem., 281, 2006
|
|
2HND
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HNY
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HNZ
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with PETT-2 | | Descriptor: | 1-[2-(4-ETHOXY-3-FLUOROPYRIDIN-2-YL)ETHYL]-3-(5-METHYLPYRIDIN-2-YL)THIOUREA, PHOSPHATE ION, Reverse transcriptase/ribonuclease H | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2GW8
 
 | | Structure of the PII signal transduction protein of Neisseria meningitidis at 1.85 resolution | | Descriptor: | PII signal transduction protein | | Authors: | Nichols, C.E, Sainsbury, S, Berrow, N.S, Alderton, D, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the P(II) signal transduction protein of Neisseria meningitidis at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1NRX
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and NAD | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-26 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1NUA
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, ZINC ION | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1NVE
 
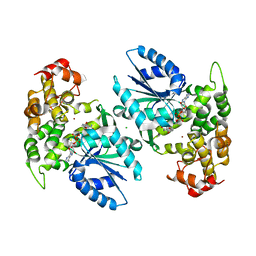 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and NAD | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1RT2
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
1NVF
 
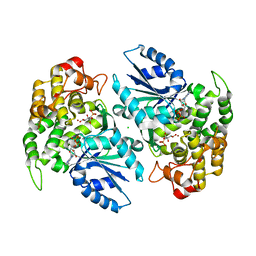 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+, ADP and carbaphosphonate | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1RT1
 
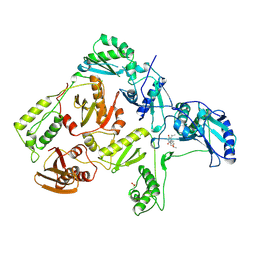 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH MKC-442 | | Descriptor: | 6-BENZYL-1-ETHOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
1RTJ
 
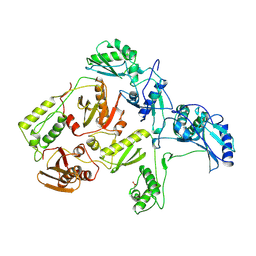 | | MECHANISM OF INHIBITION OF HIV-1 REVERSE TRANSCRIPTASE BY NON-NUCLEOSIDE INHIBITORS | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
1DRT
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND PROCLAVAMINIC ACID | | Descriptor: | 2-OXOGLUTARIC ACID, 5-AMINO-3-HYDROXY-2-(2-OXO-AZETIDIN-1-YL)-PENTANOIC ACID, CLAVAMINATE SYNTHASE 1, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DRY
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II), 2-OXOGLUTARATE AND N-ALPHA-L-ACETYL ARGININE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1REV
 
 | | HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-09-17 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design.
Structure, 3, 1995
|
|
1DS0
 
 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE | | Descriptor: | ACETATE ION, CLAVAMINATE SYNTHASE 1, SULFATE ION | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1DS1
 
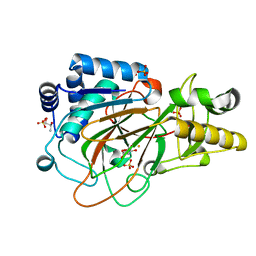 | | CRYSTAL STRUCTURE OF CLAVAMINATE SYNTHASE IN COMPLEX WITH FE(II) AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, FE (II) ION, ... | | Authors: | Zhang, Z.H, Ren, J, Stammers, D.K, Baldwin, J.E, Harlos, K, Schofield, C.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase.
Nat.Struct.Biol., 7, 2000
|
|
1OSN
 
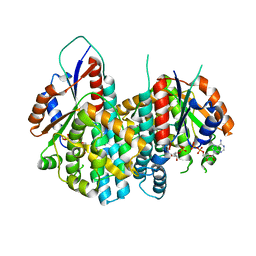 | | Crystal structure of Varicella zoster virus thymidine kinase in complex with BVDU-MP and ADP | | Descriptor: | (E)-5-(2-BROMOVINYL)-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Thymidine kinase | | Authors: | Bird, L.E, Ren, J, Wright, A, Leslie, K.D, Degreve, B, Balzarini, J, Stammers, D.K. | | Deposit date: | 2003-03-20 | | Release date: | 2003-06-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of varicella zoster virus thymidine kinase
J.Biol.Chem., 278, 2003
|
|
1TI7
 
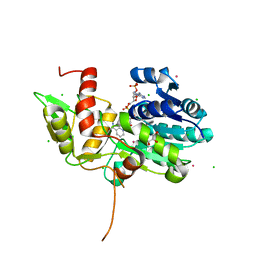 | | CRYSTAL STRUCTURE OF NMRA, A NEGATIVE TRANSCRIPTIONAL REGULATOR, IN COMPLEX WITH NADP AT 1.7A RESOLUTION | | Descriptor: | CHLORIDE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lamb, H.K, Leslie, K, Dodds, A.L, Nutley, M, Cooper, A, Johnson, C, Thompson, P, Stammers, D.K, Hawkins, A.R. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides.
J.Biol.Chem., 278, 2003
|
|
1MJT
 
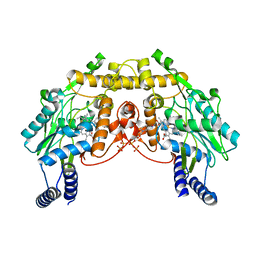 | | CRYSTAL STRUCTURE OF SANOS, A BACTERIAL NITRIC OXIDE SYNTHASE OXYGENASE PROTEIN, IN COMPLEX WITH NAD+ AND SEITU | | Descriptor: | ETHYLISOTHIOUREA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NITRIC-OXIDE SYNTHASE HOMOLOG, ... | | Authors: | Bird, L.E, Ren, J, Stammers, D.K. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of SANOS, a Bacterial Nitric Oxide Synthase Oxygenase Protein from Staphylococcus aureus
Structure, 10, 2002
|
|
1SFL
 
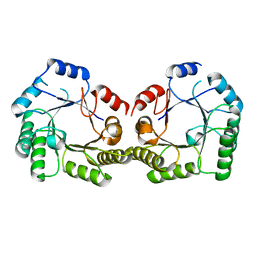 | | 1.9A Crystal structure of Staphylococcus aureus type I 3-dehydroquinase, apo form | | Descriptor: | 3-dehydroquinate dehydratase | | Authors: | Nichols, C.E, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Staphylococcus aureus type I dehydroquinase from enzyme turnover experiments.
Proteins, 56, 2004
|
|
1SFJ
 
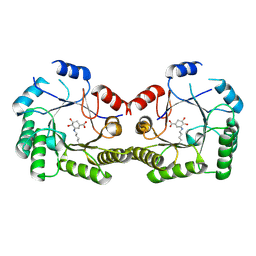 | | 2.4A Crystal structure of Staphylococcus aureus type I 3-dehydroquinase, with 3-dehydroquinate bound | | Descriptor: | 3-DEHYDROSHIKIMATE, 3-dehydroquinate dehydratase | | Authors: | Nichols, C.E, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-19 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Staphylococcus aureus type I dehydroquinase from enzyme turnover experiments
Proteins, 56, 2004
|
|
1U8A
 
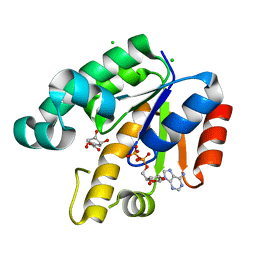 | | Crystal Structure of Mycobacterium Tuberculosis Shikimate Kinase in Complex with Shikimate and ADP at 2.15 Angstrom Resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Dhaliwal, B, Nichols, C.E, Ren, J, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-05 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic studies of shikimate binding and induced conformational changes in Mycobacterium tuberculosis shikimate kinase.
Febs Lett., 574, 2004
|
|
