2PXG
 
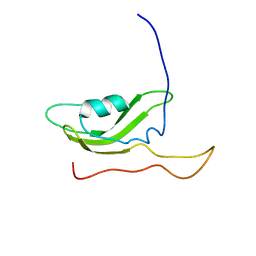 | | NMR Solution Structure of OmlA | | Descriptor: | Outer membrane protein | | Authors: | Vanini, M.M.T, Pertinhez, T.A, Sforca, M.L, Spisni, A, Benedetti, C.E. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the outer membrane lipoprotein OmlA from Xanthomonas axonopodis pv. citri reveals a protein fold implicated in protein-protein interaction.
Proteins, 71, 2008
|
|
2POA
 
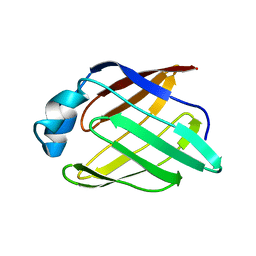 | | Schistosoma mansoni Sm14 Fatty Acid-Binding Protein: improvement of protein stability by substitution of the single Cys62 residue | | Descriptor: | 14 kDa fatty acid-binding protein | | Authors: | Ramos, C.R.R, Oyama Jr, S, Sforca, M.L, Pertinhez, T.A, Ho, P.L, Spisni, A. | | Deposit date: | 2007-04-26 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stability improvement of the fatty acid binding protein Sm14 from S. mansoni by Cys replacement: Structural and functional characterization of a vaccine candidate.
Biochim.Biophys.Acta, 1794, 2009
|
|
1DF3
 
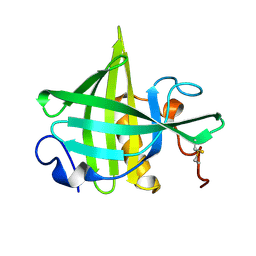 | | SOLUTION STRUCTURE OF A RECOMBINANT MOUSE MAJOR URINARY PROTEIN | | Descriptor: | MAJOR URINARY PROTEIN | | Authors: | Luecke, C, Franzoni, L, Abbate, F, Loehr, F, Ferrari, E, Sorbi, R.T, Rueterjans, H, Spisni, A. | | Deposit date: | 1999-11-17 | | Release date: | 2000-05-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant mouse major urinary protein.
Eur.J.Biochem., 266, 1999
|
|
2KQA
 
 | | The solution structure of the fungal elicitor Cerato-Platanin | | Descriptor: | Cerato-platanin | | Authors: | Oliveira, A.L, Gallo, M, Pazzagli, L, Cappugi, G, Scala, A, Cicero, D.O, Pantera, B, Spisni, A, Benedetti, C.E, Pertinhez, T.A. | | Deposit date: | 2009-11-03 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the fungal elicitor Cerato-Platanin
To be Published
|
|
2BIC
 
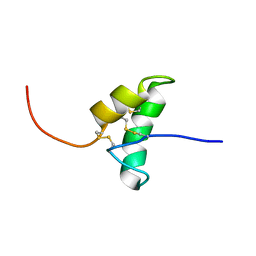 | | The solution structure of the recombinant elicitor protein PcF from the oomycete pathogen P. cactorum | | Descriptor: | PHYTOTOXIC PROTEIN PCF | | Authors: | Nicastro, G, Orsomando, G, Desario, F, Ferrari, E, Manconi, L, Spisni, A, Ruggieri, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-06-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Phytotoxic Protein Pcf: The First Characterized Member of the Phytophthora Pcf Toxin Family.
Protein Sci., 18, 2009
|
|
2JP6
 
 | | Structural and functional characterization of the recombinant form of the Kv1.3 channel blocker Tc32 | | Descriptor: | Potassium channel toxin alpha-KTx 18.1 | | Authors: | Stehling, E.G, Sforca, M.L, Zanchin, N.I, Pignatelli, A, Belluzzi, O, Spisni, A, Pertinhez, T.A. | | Deposit date: | 2007-04-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the recombinant form of the
Kv1.3 channel blocker Tc32
To be Published
|
|
2K6L
 
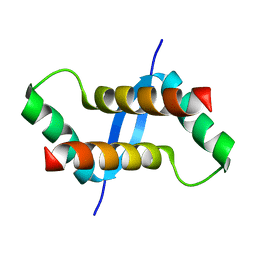 | | The solution structure of XACb0070 from Xanthonomas axonopodis pv citri reveals this new protein is a member of the RHH family of transcriptional repressors | | Descriptor: | Putative uncharacterized protein | | Authors: | Gallo, M, Cicero, D.O, Amata, I, Eliseo, T, Paci, M, Spisni, A, Ferrari, E, Pertinhez, T.A, Farah, C.S. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure reveals that XACb0070 from the plant pathogen Xanthomonas citri belongs to the RHH superfamily of bacterial DNA-binding proteins
To be Published
|
|
1JV4
 
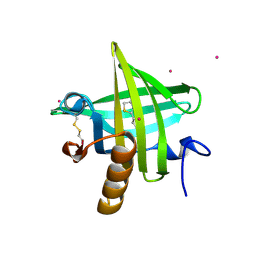 | | Crystal structure of recombinant major mouse urinary protein (rmup) at 1.75 A resolution | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, Major urinary protein 2 | | Authors: | Kuser, P.R, Franzoni, L, Ferrari, E, Spisni, A, Polikarpov, I. | | Deposit date: | 2001-08-28 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray structure of a recombinant major urinary protein at 1.75 A resolution. A comparative study of X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2F1E
 
 | | Solution structure of ApaG protein | | Descriptor: | Protein apaG | | Authors: | Contessa, G, Pertinhez, T.A, Spisni, A, Paci, M, Farah, C.S, Cicero, D.O. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ApaG from Xanthomonas axonopodis pv. citri reveals a fibronectin-3 fold.
Proteins, 67, 2007
|
|
1JBH
 
 | | Solution structure of cellular retinol binding protein type-I in the ligand-free state | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-06-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of Apo- and holo-cellular retinol-binding protein in solution.
J.Biol.Chem., 277, 2002
|
|
1KGL
 
 | | Solution structure of cellular retinol binding protein type-I in complex with all-trans-retinol | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I, RETINOL | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-11-27 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Backbone Dynamics of Apo- and Holo-cellular Retinol-binding
Protein in Solution.
J.Biol.Chem., 277, 2002
|
|
1H5O
 
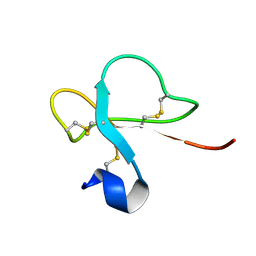 | | Solution structure of Crotamine, a neurotoxin from Crotalus durissus terrificus | | Descriptor: | MYOTOXIN | | Authors: | Nicastro, G, Franzoni, L, De Chiara, C, Mancin, C.A, Giglio, J.R, Spisni, A. | | Deposit date: | 2001-05-23 | | Release date: | 2003-05-09 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Crotamine, a Na+ Channel Affecting Toxin from Crotalus Durissus Terrificus Venom
Eur.J.Biochem., 270, 2003
|
|
