4MV5
 
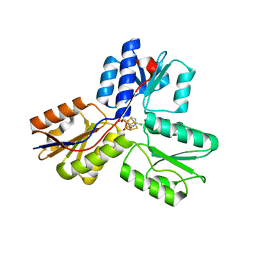 | | IspH in complex with 6-chloropyridin-3-ylmethyl diphosphate | | Descriptor: | (6-chloropyridin-3-yl)methyl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
4XS3
 
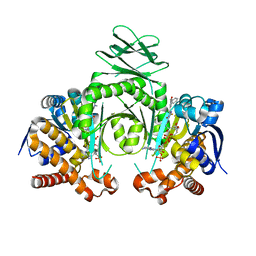 | | Crystal structure of a metabolic reductase with (E)-1-benzyl-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one | | Descriptor: | (E)-1-benzyl-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, B, Wu, F, Jiang, H, Kogiso, M, Yao, Y, Zhou, C, Li, X, Song, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.291 Å) | | Cite: | Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds.
J.Med.Chem., 58, 2015
|
|
5VBL
 
 | | Structure of apelin receptor in complex with agonist peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor,Rubredoxin,Apelin receptor Chimera, ZINC ION, ... | | Authors: | Ma, Y, Yue, Y, Ma, Y, Zhang, Q, Zhou, Q, Song, Y, Shen, Y, Li, X, Ma, X, Li, C, Hanson, M.A, Han, G.W, Sickmier, E.A, Swaminath, G, Zhao, S, Stevems, R.C, Hu, L.A, Zhong, W, Zhang, M, Xu, F. | | Deposit date: | 2017-03-29 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Apelin Control of the Human Apelin Receptor
Structure, 25, 2017
|
|
9JJH
 
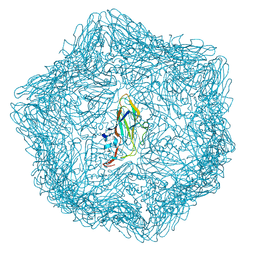 | | Cryo-EM structure of a T=1 VLP of RHDV GI.2 with N-terminal 1-37 residues truncated | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP
J.Virol.
|
|
9JJJ
 
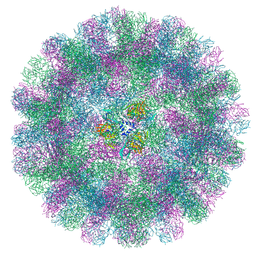 | | Cryo-EM structure of a T=3 VLP of RHDV GI.2 | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP
J.Virol.
|
|
9JJG
 
 | | Cryo-EM structure of RHDV GI.2 virion | | Descriptor: | Genome polyprotein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP
J.Virol., 2014
|
|
9JJI
 
 | | Local refinement of RHDV GI.2 T=1 VLP | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP
J.Virol.
|
|
6MO1
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 5-[4-(aminomethyl)phenyl]-6-[4-(furan-3-yl)phenyl]-N-[(piperidin-4-yl)methyl]pyrazin-2-amine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Hua, Y, Nie, S, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
6MO0
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{3-[4-(furan-3-yl)phenyl]-5-[(piperidin-4-yl)methoxy]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J. Am. Chem. Soc., 141, 2019
|
|
6MO2
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{5-[(piperidin-4-yl)methoxy]-3-[4-(1H-pyrazol-4-yl)phenyl]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
8DH6
 
 | | Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cytochrome c oxidase subunit 1, ... | | Authors: | Godoy, A.S, Song, Y, Cheruvara, H, Quigley, A, Oliva, G. | | Deposit date: | 2022-06-25 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs
To Be Published
|
|
8DH7
 
 | |
5V58
 
 | |
5V59
 
 | |
8TPE
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPG
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPF
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPB
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-2-chloroacetamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPH
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPI
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-2-hydroxy-2-methylpropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPD
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | 3C-like proteinase nsp5, N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[3-(2-chloroacetamido)phenyl]furan-2-carboxamide | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPC
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[4-(2-chloroacetamido)phenyl]furan-2-carboxamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
9IUZ
 
 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
4S0J
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123V mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
