5KZM
 
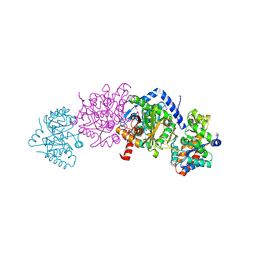 | | Crystal structure of Tryptophan synthase alpha-beta chain complex from Francisella tularensis | | Descriptor: | ACETATE ION, CALCIUM ION, Tryptophan synthase alpha chain, ... | | Authors: | Chang, C, Michalska, K, Joachimiak, G, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-10 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Conservation of the structure and function of bacterial tryptophan synthases.
Iucrj, 6, 2019
|
|
4WD8
 
 | |
4QMK
 
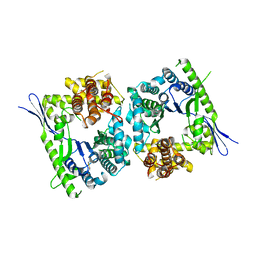 | | Crystal structure of type III effector protein ExoU (exoU) | | Descriptor: | BETA-MERCAPTOETHANOL, Type III secretion system effector protein ExoU | | Authors: | Halavaty, A.S, Tyson, G.H, Zhang, A, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-16 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Phosphatidylinositol 4,5-Bisphosphate Binding Domain Mediates Plasma Membrane Localization of ExoU and Other Patatin-like Phospholipases.
J.Biol.Chem., 290, 2015
|
|
4WB7
 
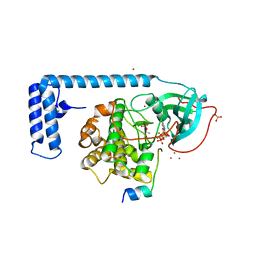 | | Crystal structure of a chimeric fusion of human DnaJ (Hsp40) and cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R3C
 
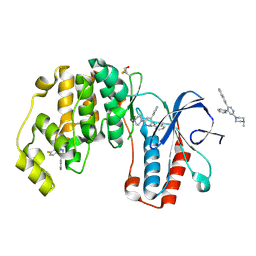 | | Crystal structure of p38 alpha MAP kinase in complex with a novel isoform selective drug candidate | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, 6-(4-methylpiperazin-1-yl)-3-(naphthalen-2-yl)-4-(pyridin-4-yl)pyridazine, CHLORIDE ION, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Targeting human central nervous system protein kinases: An isoform selective p38 alpha MAPK inhibitor that attenuates disease progression in Alzheimer's disease mouse models.
ACS Chem Neurosci, 6, 2015
|
|
4U9D
 
 | |
4U9E
 
 | |
4WB8
 
 | | Crystal structure of human cAMP-dependent protein kinase A (catalytic alpha subunit), exon 1 deletion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4JRR
 
 | | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Shumilin, I.A, Jameson-Lee, M, Cymborowski, M, Domagalski, M.J, Chertihin, O, Kpadeh, Z.Z, Yeh, A.J, Hoffman, P.S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila
To be Published
|
|
5HPO
 
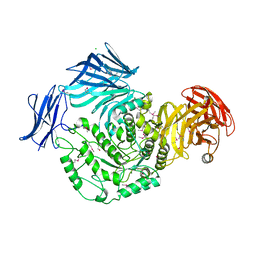 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with maltopentaose | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Winsor, J, Grimshaw, S, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5HXM
 
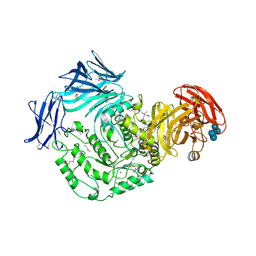 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with panose | | Descriptor: | Alpha-xylosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Winsor, J, Grimshaw, S, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-31 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
8UTG
 
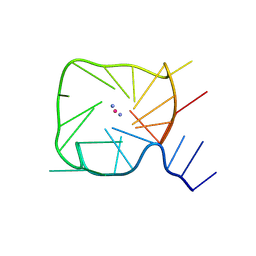 | | An RNA G-Quadruplex from the NS5 gene in the West Nile Virus Genome | | Descriptor: | AMMONIUM ION, POTASSIUM ION, RNA (5'-R(*UP*AP*UP*GP*GP*AP*AP*GP*AP*GP*GP*CP*GP*GP*UP*UP*GP*GP*UP*AP*U)-3') | | Authors: | Terrell, J.R, Le, T.T, Wilson, W.D, Siemer, J.L. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of an RNA G-quadruplex from the West Nile virus genome.
Nat Commun, 15, 2024
|
|
5JQA
 
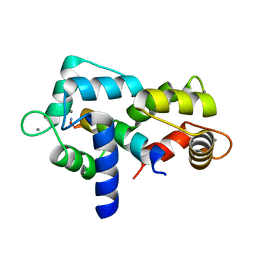 | | CaM:RM20 complex | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase, ... | | Authors: | Tokars, V.L, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2016-05-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CaM:RM20 complex
To be Published
|
|
3FF1
 
 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
5JTH
 
 | | Crystal structure of E67A calmodulin - CaM:RM20 analog complex | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2016-05-09 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of E67A calmodulin - CaM:RM20 analog complex
To Be Published
|
|
4EGU
 
 | | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ZINC ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile
To be Published
|
|
3NX4
 
 | | Crystal structure of the yhdH oxidoreductase from Salmonella enterica in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Peterson, S, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-12 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the yhdH oxidoreductase from Salmonella enterica in complex with NADP
To be Published
|
|
8T4N
 
 | |
7GLC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6f6ae286-3 (Mpro-P1858) | | Descriptor: | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-4-methoxy-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GLU
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6479a3a9-2 (Mpro-P2028) | | Descriptor: | 2-(5-chloro-2-{[(methanesulfonyl)amino]methyl}phenyl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.627 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GM9
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-1bed62cf-3 (Mpro-P2113) | | Descriptor: | 2-[(1'P,3'S)-6-chloro-1'-(isoquinolin-4-yl)-2',5'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylacetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GMO
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-12c4873b-5 (Mpro-P2206) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-2-[(3R)-2-oxopyrrolidin-3-yl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GLS
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-bfb445d4-2 (Mpro-P2011) | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GM8
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-7889e8da-3 (Mpro-P2101) | | Descriptor: | (4S)-6-chloro-N-(7-chloroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GN6
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e119ab4f-5 (Mpro-P2358) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-4-methyl-2-[2-(methylamino)-2-oxoethyl]-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
