6TAT
 
 | |
3GEU
 
 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
6TAP
 
 | | Structure of the dArc1 capsid | | Descriptor: | Activity-regulated cytoskeleton associated protein 1, ZINC ION | | Authors: | Erlendsson, S, Morado, D.R, Shepherd, J.D, Briggs, J.A.G. | | Deposit date: | 2019-10-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins.
Nat.Neurosci., 23, 2020
|
|
6TAS
 
 | | Structure of the two-fold capsomer of the dArc1 capsid | | Descriptor: | Activity-regulated cytoskeleton associated protein 1, ZINC ION | | Authors: | Erlendsson, S, Morado, D.R, Shepherd, J.D, Briggs, J.A.G. | | Deposit date: | 2019-10-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins.
Nat.Neurosci., 23, 2020
|
|
2WKF
 
 | | Crystal Structure of Macrophage Migration Inhibitory Factor from Plasmodium falciparum | | Descriptor: | GLYCEROL, MACROPHAGE MIGRATION INHIBITORY FACTOR | | Authors: | Dobson, S.E, Augustijn, K.D, Brannigan, J.A, Dodson, E.J, Waters, A.P, Wilkinson, A.J. | | Deposit date: | 2009-06-11 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structures of macrophage migration inhibitory factor from Plasmodium falciparum and Plasmodium berghei.
Protein Sci., 18, 2009
|
|
4MHB
 
 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
1OTE
 
 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P65 AT 110K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, photoactive yellow protein, PYP | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTA
 
 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4AYE
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 E283AE304A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYI
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 Wild type | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, LIPOPROTEIN GNA1870 CCOMPND 7 | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYN
 
 | | Structure of the C-terminal barrel of Neisseria meningitidis FHbp Variant 2 | | Descriptor: | FACTOR H-BINDING PROTEIN, SULFATE ION | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
8JBO
 
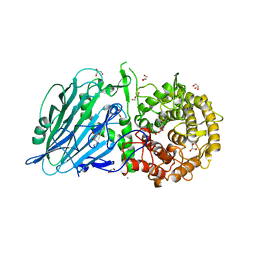 | | Crystal structure of TxGH116 from Thermoanaerobacterium xylanolyticum with isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2023-05-09 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of a GH116 beta-glucosidase and its missense mutants by GBA2 inhibitors: Crystallographic and quantum chemical study.
Chem.Biol.Interact., 384, 2023
|
|
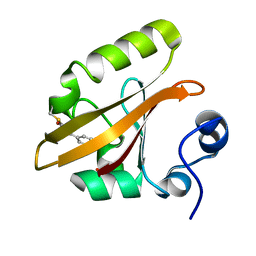
1OT6
 
 | |
4AYM
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 P106A mutant | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
8UPL
 
 | | Cryo-EM structure of a Clockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
1OT9
 
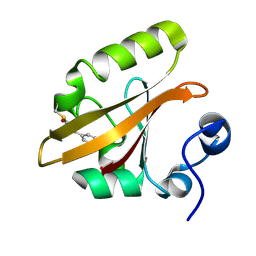 | | CRYOTRAPPED STATE IN WILD TYPE PHOTOACTIVE YELLOW PROTEIN, INDUCED WITH CONTINUOUS ILLUMINATION AT 110K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTI
 
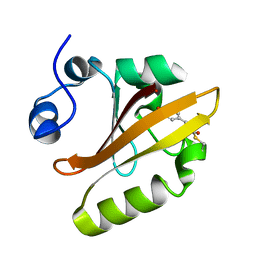 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P65 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6ETI
 
 | | Structure of inhibitor-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Altmann, K.H, Locher, K.P. | | Deposit date: | 2017-10-26 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FDF
 
 | |
8UOX
 
 | | Cryo-EM structure of a Counterclockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UMD
 
 | | Cryo-EM structure of a single subunit of a Counterclockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring. | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-17 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UMX
 
 | | Cryo-EM structure of a single subunit of a Clockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring. | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
7Z9P
 
 | | The novel DNA binding mechanism of ridinilazole, a precision Clostridiodes difficile antibiotic | | Descriptor: | DNA (5'-D(P*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G*(RID))-3'), Ridinilazole, SODIUM ION, ... | | Authors: | Mason, S, Leonard, P.M. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Novel DNA Binding Mechanism of Ridinilazole, a Precision Clostridiodes difficile Antibiotic.
Antimicrob.Agents Chemother., 67, 2023
|
|
1OTB
 
 | | WILD TYPE PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
