6QQN
 
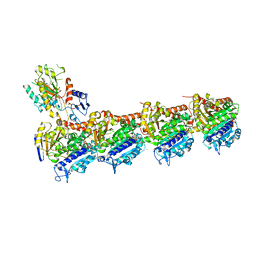 | | Tubulin-TH588 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Patterson, J.C, Joughin, B.A, Prota, A.E, Muehlethaler, T, Jonas, O.H, Whitman, M.A, Varmeh, S, Chen, S, Balk, S.P, Steinmetz, M.O, Lauffenburger, D.A, Yaffe, M.B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | VISAGE Reveals a Targetable Mitotic Spindle Vulnerability in Cancer Cells.
Cell Syst, 9, 2019
|
|
2Y3R
 
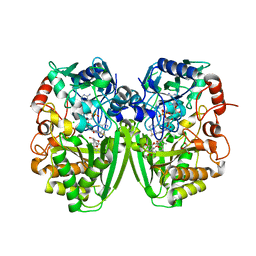 | | Structure of the tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P21 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y3S
 
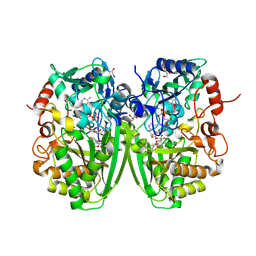 | | Structure of the tirandamycine-bound FAD-dependent tirandamycin oxidase TamL in C2 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y08
 
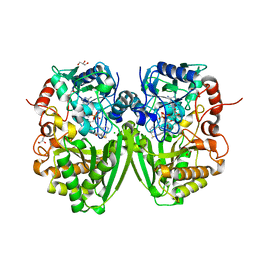 | | Structure of the substrate-free FAD-dependent tirandamycin oxidase TamL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-11-30 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y4G
 
 | | Structure of the Tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P212121 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2011-01-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
1GG8
 
 | | DESIGN OF INHIBITORS OF GLYCOGEN PHOSPHORYLASE: A STUDY OF ALPHA-AND BETA-C-GLUCOSIDES AND 1-THIO-BETA-D-GLUCOSE COMPOUNDS | | Descriptor: | ALPHA-D-GLUCOPYRANOSYL-2-CARBOXYLIC ACID AMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Watson, K.A, Mitchell, E.P, Johnson, L.N, Son, J.C, Bichard, C.J, Orchard, M.G, Fleet, G.W, Oikonomakos, N.G, Leonidas, D.D, Kontou, M, Papageorgiou, A.C. | | Deposit date: | 2000-07-30 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design of inhibitors of glycogen phosphorylase: a study of alpha- and beta-C-glucosides and 1-thio-beta-D-glucose compounds.
Biochemistry, 33, 1994
|
|
2GDM
 
 | | LEGHEMOGLOBIN (OXY) | | Descriptor: | LEGHEMOGLOBIN (OXY), OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Harutyunyan, E.H, Safonova, T.N, Kuranova, I.P, Popov, A.N, Teplyakov, A.V, Obmolova, G.V, Rusakov, A.A, Dodson, G.G, Wilson, J.C, Perutz, M.F. | | Deposit date: | 1994-09-14 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Deoxy-and Oxy-Leghaemoglobin from Lupin
J.Mol.Biol., 251, 1995
|
|
2VK7
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, EXO-ALPHA-SIALIDASE | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VK5
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | CALCIUM ION, EXO-ALPHA-SIALIDASE, GLYCEROL | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2VK6
 
 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, EXO-ALPHA-SIALIDASE, ... | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
2W38
 
 | | Crystal structure of the pseudaminidase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SIALIDASE | | Authors: | Xu, G, Ryan, C, Kiefel, M.J, Wilson, J.C, Taylor, G.L. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on the Pseudomonas Aeruginosa Sialidase-Like Enzyme Pa2794 Suggest Substrate and Mechanistic Variations.
J.Mol.Biol., 386, 2009
|
|
6HPS
 
 | | Near-infrared dual bioluminescence imaging in vivo using infra-luciferin | | Descriptor: | Luciferin 4-monooxygenase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[[2-[(~{E})-2-(6-oxidanyl-1,3-benzothiazol-2-yl)ethenyl]-1,3-thiazol-4-yl]carbonyl]sulfamate | | Authors: | Parkinson, G.N, Stowe, C, Anderson, J.C. | | Deposit date: | 2018-09-21 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Near-infrared dual bioluminescence imaging in mouse models of cancer using infraluciferin.
Elife, 8, 2019
|
|
4MUB
 
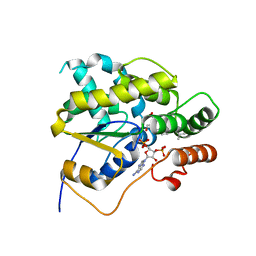 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/Oxamniquine Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase, {(2S)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Valentim, C.L.L, Cioli, D, Chevalier, F.D, Cao, X, Taylor, A.B, Holloway, S.P, Pica-Mattoccia, L, Guidi, A, Basso, A, Tsai, I.J, Berriman, M, Carvalho-Queiroz, C, Almeida, M, Aguilar, H, Frantz, D.E, Hart, P.J, Anderson, T.J.C, LoVerde, P.T. | | Deposit date: | 2013-09-21 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and molecular basis of drug resistance and species-specific drug action in schistosome parasites.
Science, 342, 2013
|
|
4MUA
 
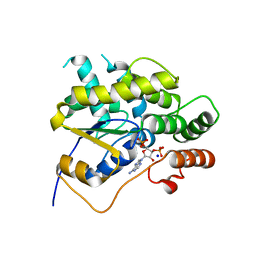 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, Sulfotransferase | | Authors: | Valentim, C.L.L, Cioli, D, Chevalier, F.D, Cao, X, Taylor, A.B, Holloway, S.P, Pica-Mattoccia, L, Guidi, A, Basso, A, Tsai, I.J, Berriman, M, Carvalho-Queiroz, C, Almeida, M, Aguilar, H, Frantz, D.E, Hart, P.J, Anderson, T.J.C, LoVerde, P.T. | | Deposit date: | 2013-09-21 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetic and molecular basis of drug resistance and species-specific drug action in schistosome parasites.
Science, 342, 2013
|
|
1W0P
 
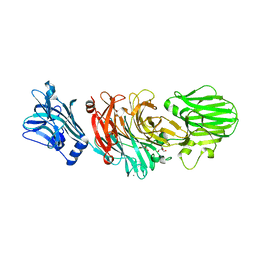 | | Vibrio cholerae sialidase with alpha-2,6-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Moustafa, I, Connaris, H, Taylor, M, Zaitsev, V, Wilson, J.C, Kiefel, M.J, von-Itzstein, M, Taylor, G. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sialic Acid Recognition by Vibrio Cholerae Neuraminidase.
J.Biol.Chem., 279, 2004
|
|
1AOK
 
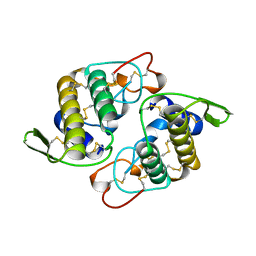 | | VIPOXIN COMPLEX | | Descriptor: | ACETATE ION, VIPOXIN COMPLEX | | Authors: | Perbandt, M, Wilson, J.C, Eschenburg, S, Betzel, C. | | Deposit date: | 1997-07-07 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of vipoxin at 2.0 A: an example of regulation of a toxic function generated by molecular evolution.
FEBS Lett., 412, 1997
|
|
1W0O
 
 | | Vibrio cholerae sialidase | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Moustafa, I, Connaris, H, Taylor, M, Zaitsev, V, Wilson, J.C, Kiefel, M.J, von-Itzstein, M, Taylor, G. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sialic Acid Recognition by Vibrio Cholerae Neuraminidase
J.Biol.Chem., 279, 2004
|
|
2F13
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor | | Descriptor: | (2R)-2,3-dihydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor
To be Published
|
|
2F11
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with isobutyl ether mimetic Inhibitor | | Descriptor: | 2-methylpropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with isobutyl ether mimetic Inhibitor
To be Published
|
|
4GB1
 
 | | Synthesis and Evaluation of Novel 3-C-alkylated-Neu5Ac2en Derivatives as Probes of Influenza Virus Sialidase 150-loop flexibility | | Descriptor: | 5-acetamido-2,6-anhydro-3,5-dideoxy-3-[(2E)-3-phenylprop-2-en-1-yl]-D-glycero-L-altro-non-2-enonic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Rudrawar, S, Rameix-Welti, M.-A, Maggioni, A, Dyason, J.C, Rose, F.J, van der Werf, S, Thomson, R.J, Naffakh, N, Russell, R.J.M, von Itzstein, M. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Synthesis and evaluation of novel 3-C-alkylated-Neu5Ac2en derivatives as probes of influenza virus sialidase 150-loop flexibility.
Org.Biomol.Chem., 10, 2012
|
|
2F12
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor | | Descriptor: | 3-hydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor
To be Published
|
|
7QU6
 
 | | Crystal structure of the N-terminal domain of Siglec-8 | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Atxabal, U, Nycholat, C.M, Oyenarte, I, Paulson, J.C, Franconetti, A, Quintana, J.I, Unione, L, Delgado, S, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
1SDO
 
 | | Crystal Structure of Restriction Endonuclease BstYI | | Descriptor: | BstYI | | Authors: | Townson, S.A, Samuelson, J.C, Vanamee, E.S, Edwards, T.A, Escalante, C.R, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2004-02-13 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of BstYI at 1.85 A Resolution: A Thermophilic Restriction Endonuclease with Overlapping Specificities to BamHI and BglII
J.Mol.Biol., 338, 2004
|
|
1VRR
 
 | | Crystal structure of the restriction endonuclease BstYI complex with DNA | | Descriptor: | 5'-D(*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3', BstYI | | Authors: | Townson, S.A, Samuelson, J.C, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2005-06-02 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Implications for Switching Restriction Enzyme Specificities from the Structure of BstYI Bound to a BglII DNA Sequence.
Structure, 13, 2005
|
|
1L3L
 
 | | Crystal structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*GP*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP*CP*TP*GP*CP*AP*CP*AP*TP*C)-3', Transcriptional activator protein traR | | Authors: | Zhang, R, Pappas, T, Brace, J.L, Miller, P.C, Oulmassov, T, Molyneaux, J.M, Anderson, J.C, Bashkin, J.K, Winans, S.C, Joachimiak, A. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA.
Nature, 417, 2002
|
|
