3PT6
 
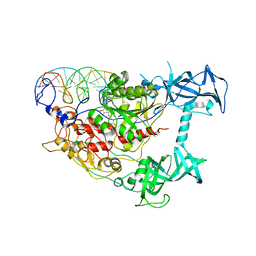 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
7EKR
 
 | |
2W0D
 
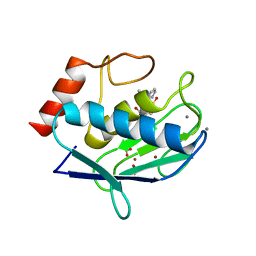 | | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? A Case Study of Metalloproteinases. | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Isaksson, J, Nystrom, S, Derbyshire, D.J, Wallberg, H, Agback, T, Kovacs, H, Bertini, I, Felli, I.C. | | Deposit date: | 2008-08-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? a Case Study of Metalloproteinases.
J.Med.Chem., 52, 2009
|
|
3MBZ
 
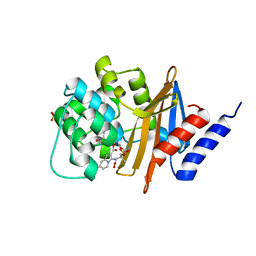 | | OXA-24 beta-lactamase complex soaked with 10mM SA4-17 inhibitor for 15min | | Descriptor: | (2S,3R)-2-[(7-aminocarbonyl-2-methanoyl-indolizin-3-yl)amino]-4-aminocarbonyloxy-3-methyl-3-sulfino-butanoic acid, Betalactamase OXA24, SULFATE ION | | Authors: | Sampson, J, van den Akker, F. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase
J.Am.Chem.Soc., 132, 2010
|
|
3BZM
 
 | | Crystal Structure of Open form of Menaquinone-Specific Isochorismate Synthase, MenF | | Descriptor: | CITRIC ACID, Menaquinone-specific isochorismate synthase | | Authors: | Parsons, J.F, Shi, K.M, Ladner, J.E. | | Deposit date: | 2008-01-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of isochorismate synthase in complex with magnesium.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3Q3N
 
 | | Toluene 4 monooxygenase HD complex with p-nitrophenol | | Descriptor: | FE (III) ION, P-NITROPHENOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Acheson, J.F, Bailey, L.J, Fox, B.G. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
3Q2A
 
 | | Toluene 4 monooxygenase HD complex with inhibitor p-aminobenzoate | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-AMINOBENZOIC ACID, FE (III) ION, ... | | Authors: | Acheson, J.F, Bailey, L.J, Fox, B.G. | | Deposit date: | 2010-12-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
3Q3O
 
 | | Toluene 4 monooxygenase HD complex with phenol | | Descriptor: | FE (III) ION, PENTAETHYLENE GLYCOL, PHENOL, ... | | Authors: | Acheson, J.F, Bailey, L.J, Fox, B.G. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
3BRF
 
 | | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, C2221 | | Descriptor: | DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), Lin-12 and glp-1 phenotype protein 1, ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
8SH7
 
 | |
6B9M
 
 | |
1R31
 
 | | HMG-CoA reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, COENZYME A, ... | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-09-30 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-CoA Reductase.
To be Published
|
|
6B9L
 
 | | Crystal structure of EphA2 with peptide 135E2 | | Descriptor: | Ephrin type-A receptor 2, HIS TAG CLEAVED OFF, peptide 135E2, ... | | Authors: | Song, J, Tan, X. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Based Design of Novel EphA2 Agonistic Agents with Nanomolar Affinity in Vitro and in Cell.
ACS Chem. Biol., 13, 2018
|
|
1R7I
 
 | | HMG-CoA Reductase from P. mevalonii, native structure at 2.2 angstroms resolution. | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, SULFATE ION | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-10-21 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-COA Reductase.
To be Published
|
|
8U1C
 
 | |
8U1Q
 
 | |
3PTA
 
 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
2Y3R
 
 | | Structure of the tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P21 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y3S
 
 | | Structure of the tirandamycine-bound FAD-dependent tirandamycin oxidase TamL in C2 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y08
 
 | | Structure of the substrate-free FAD-dependent tirandamycin oxidase TamL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-11-30 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y4G
 
 | | Structure of the Tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P212121 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2011-01-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2MZW
 
 | | Staphylococcus aureus FusB:EF-GC3 complex | | Descriptor: | Elongation factor G, Far1, ZINC ION | | Authors: | Tomlinson, J.H, Thompson, G.S, Kalverda, A.P, Zhuravleva, A, O'Neill, A. | | Deposit date: | 2015-02-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A target-protection mechanism of antibiotic resistance at atomic resolution: insights into FusB-type fusidic acid resistance.
Sci Rep, 6, 2016
|
|
4R79
 
 | |
3OPP
 
 | | ESBL R164S mutant of SHV-1 beta-lactamase complexed with SA2-13 | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Sampson, J.M, van den Akker, F. | | Deposit date: | 2010-09-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand-dependent disorder of the Omega loop observed in extended-spectrum SHV-type beta-lactamase.
Antimicrob.Agents Chemother., 55, 2011
|
|
7M16
 
 | | Triazole-based BET family bromodomain inhibitor bound to BRD4(D1) | | Descriptor: | 4-{5-[6-(3,5-dimethylanilino)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidine-1-carboximidamide, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W. | | Deposit date: | 2021-03-12 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Systematically Mitigating the p38alpha Activity of Triazole-based BET Inhibitors
ACS Med. Chem. Lett., 10, 2019
|
|
