3K9Z
 
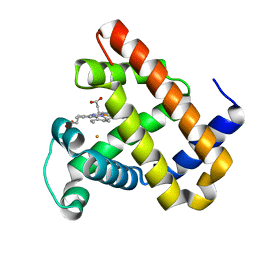 | | Rational Design of a Structural and Functional Nitric Oxide Reductase | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yeung, N, Lin, Y.-W, Gao, Y.-G, Zhao, X, Russell, B.S, Lei, L, Miner, K.D, Robinson, H, Lu, Y. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of a structural and functional nitric oxide reductase.
Nature, 462, 2009
|
|
3CS1
 
 | | Flagellar Calcium-binding Protein (FCaBP) from T. cruzi | | Descriptor: | Flagellar calcium-binding protein | | Authors: | Ames, J.B, Ladner, J.E, Wingard, J.N, Robinson, H, Fisher, A. | | Deposit date: | 2008-04-08 | | Release date: | 2008-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Membrane Targeting by the Flagellar Calcium-binding Protein (FCaBP), a Myristoylated and Palmitoylated Calcium Sensor in Trypanosoma cruzi.
J.Biol.Chem., 283, 2008
|
|
3MCA
 
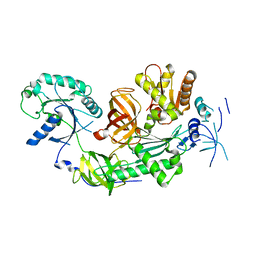 | |
5V8D
 
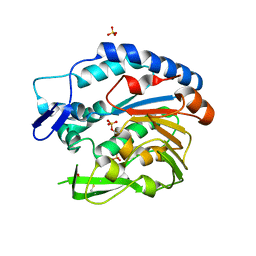 | | Structure of Bacillus cereus PatB1 with sulfonyl adduct | | Descriptor: | Bacillus cereus PatB1, SULFATE ION | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
5V1Z
 
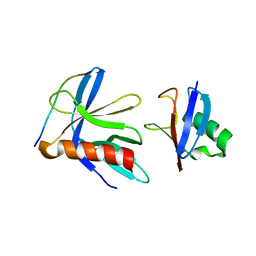 | | Crystal structure of the RPN13 PRU-RPN2 (932-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
5V1Y
 
 | | Crystal structure of the ternary RPN13 PRU-RPN2 (940-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
3M38
 
 | | The roles of Glutamates and Metal ions in a rationally designed nitric oxide reductase based on myoglobin: I107E FeBMb (No metal ion binding to FeB site) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | Deposit date: | 2010-03-08 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M3B
 
 | | The roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin: Zn(II)-I107E FeBMb (Zn(II) binding to FeB site) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | Deposit date: | 2010-03-08 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5WZ1
 
 | | Crystal structure of Zika virus NS5 methyltransferase bound to S-adenosyl-L-methionine | | Descriptor: | NS5 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
3MN0
 
 | | Introducing a 2-His-1-Glu Non-Heme Iron Center into Myoglobin confers Nitric Oxide Reductase activity: Cu(II)-CN-FeBMb(-His) form | | Descriptor: | COPPER (II) ION, CYANIDE ION, Myoglobin, ... | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Lei, L, Robinson, H, Lu, Y. | | Deposit date: | 2010-04-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Introducing a 2-his-1-glu nonheme iron center into myoglobin confers nitric oxide reductase activity.
J.Am.Chem.Soc., 132, 2010
|
|
2QKL
 
 | |
4H6J
 
 | | Identification of Cys 255 in HIF-1 as a novel site for development of covalent inhibitors of HIF-1 /ARNT PasB domain protein-protein interaction. | | Descriptor: | ARYL HYDROCARBON NUCLEAR TRANSLOCATOR, HYPOXIA INDUCIBLE FACTOR 1-ALPHA | | Authors: | Cardoso, R, Love, R.A, Nilsson, C, Bergqvist, S, Nowlin, D, Yan, J, Liu, K, Zhu, J, Chen, P, Deng, Y.-L, Dyson, H.J, Greig, M.J, Brooun, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Identification of Cys255 in HIF-1 alpha as a novel site for development of covalent inhibitors of HIF-1 alpha /ARNT PasB domain protein-protein interaction.
Protein Sci., 21, 2012
|
|
5WZ2
 
 | | Crystal structure of Zika virus NS5 methyltransferase bound to SAM and RNA analogue (m7GpppA) | | Descriptor: | NS5 MTase, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
5WZ3
 
 | | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase(RdRP) | | Descriptor: | NS5 RdRp, ZINC ION | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
5TCB
 
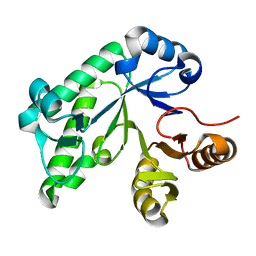 | |
2ZJG
 
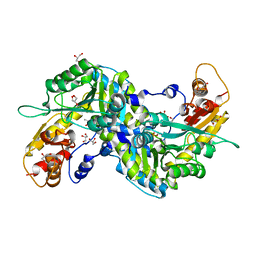 | | Crystal structural of mouse kynurenine aminotransferase III | | Descriptor: | GLYCEROL, Kynurenine-oxoglutarate transaminase 3 | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-03-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of mouse kynurenine aminotransferase III
To be Published
|
|
2R5E
 
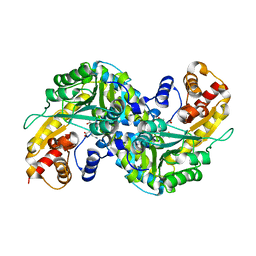 | | Aedes kynurenine aminotransferase in complex with glutamine | | Descriptor: | Kynurenine aminotransferase, N~2~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-GLUTAMINE | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Li, J. | | Deposit date: | 2007-09-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
Biochemistry, 47, 2008
|
|
3NIH
 
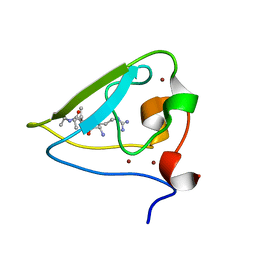 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3EVR
 
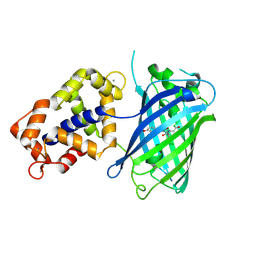 | | Crystal structure of Calcium bound monomeric GCAMP2 | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
2HTS
 
 | |
3EVP
 
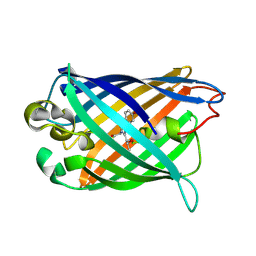 | | crystal structure of circular-permutated EGFP | | Descriptor: | Green fluorescent protein,Green fluorescent protein | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
2IP2
 
 | |
5U9B
 
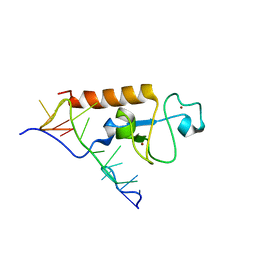 | | Solution structure of the zinc fingers 1 and 2 of MBNL1 in complex with human cardiac troponin T pre-mRNA | | Descriptor: | Muscleblind-like protein 1, RNA (5'-R(P*GP*UP*CP*UP*CP*GP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
5U6L
 
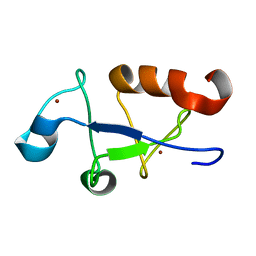 | | Solution structure of the zinc fingers 3 and 4 of MBNL1 | | Descriptor: | Muscleblind-like protein 1, ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
4OWW
 
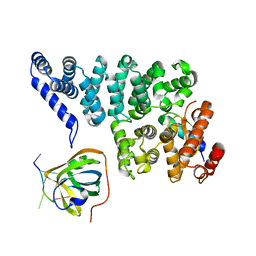 | | Structural basis of SOSS1 in complex with a 35nt ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), Integrator complex subunit 3, SOSS complex subunit B1, ... | | Authors: | Ren, W, Sun, Q, Tang, X, Song, H. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Cell Rep, 6, 2014
|
|
