2OEX
 
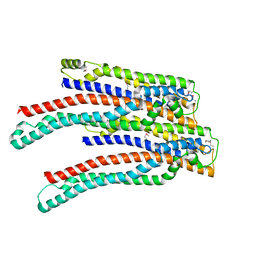 | | Structure of ALIX/AIP1 V Domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2OGD
 
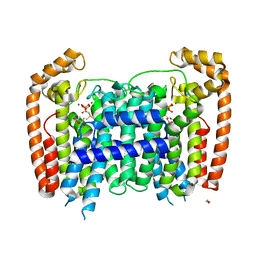 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-527 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2007-01-05 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisphosphonates: Teaching Old Drugs with New Tricks
TO BE PUBLISHED
|
|
2O6W
 
 | | Crystal Structure of a Pentapeptide Repeat Protein (Rfr23) from the cyanobacterium Cyanothece 51142 | | Descriptor: | ARSENIC, Repeat Five Residue (Rfr) protein or pentapeptide repeat protein | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H, Pakrasi, H.B. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the structural variation between pentapeptide repeat proteins-Crystal structure of Rfr23 from Cyanothece 51142.
J.Struct.Biol., 162, 2008
|
|
2OEV
 
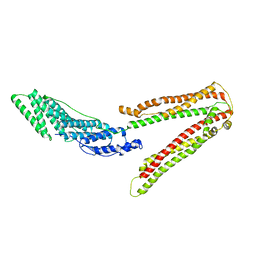 | | Crystal structure of ALIX/AIP1 | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
1Q67
 
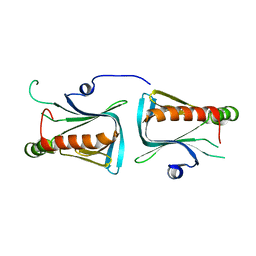 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
2KA4
 
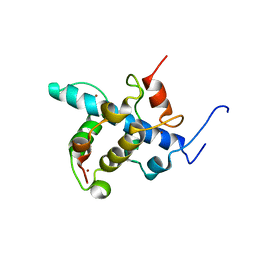 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
1G7O
 
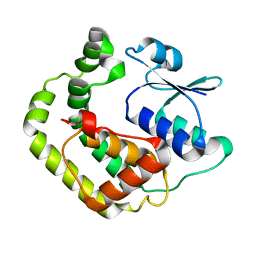 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | Descriptor: | GLUTAREDOXIN 2 | | Authors: | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-11-10 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
1FBQ
 
 | |
1R8U
 
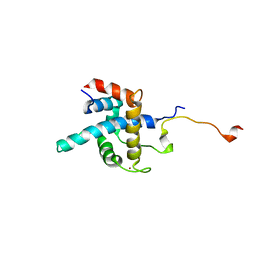 | | NMR structure of CBP TAZ1/CITED2 complex | | Descriptor: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | Authors: | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
1PN9
 
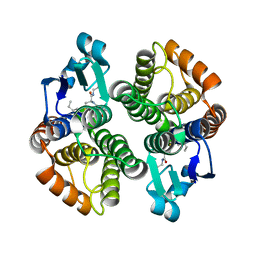 | | Crystal structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae | | Descriptor: | Glutathione S-transferase 1-6, S-HEXYLGLUTATHIONE | | Authors: | Chen, L, Hall, P.R, Zhou, X.E, Ranson, H, Hemingway, J, Meehan, E.J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1S4T
 
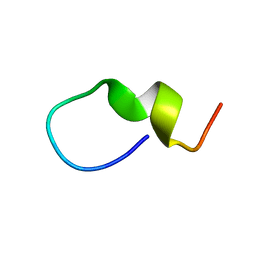 | | Solution structure of synthetic 21mer peptide spanning region 135-155 (in human numbering) of sheep prion protein | | Descriptor: | Major prion protein | | Authors: | Kozin, S.A, Lepage, C, Hui Bon Hoa, G, Rabesona, H, Mazur, A.K, Blond, A, Cheminant, M, Haertle, T, Debey, P, Rebuffat, S. | | Deposit date: | 2004-01-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific recognition between surface loop 2 (132-143) and helix 1 (144-154)
within sheep prion protein from in vitro studies of synthetic peptides
To be Published
|
|
1S2M
 
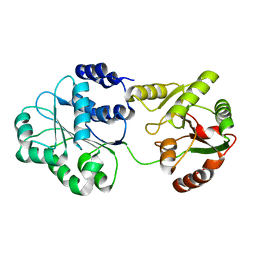 | |
1RXR
 
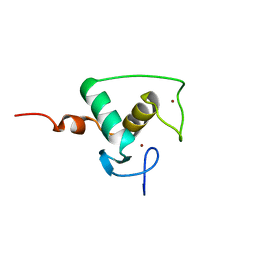 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE RETINOID X RECEPTOR DNA BINDING DOMAIN, NMR, 20 STRUCTURE | | Descriptor: | RETINOIC ACID RECEPTOR-ALPHA, ZINC ION | | Authors: | Holmbeck, S.M.A, Foster, M.P, Casimiro, D.R, Sem, D.S, Dyson, H.J, Wright, P.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the retinoid X receptor DNA-binding domain.
J.Mol.Biol., 281, 1998
|
|
1SB0
 
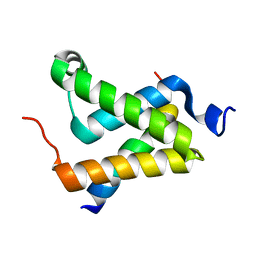 | | Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb | | Descriptor: | protein CBP, protein c-Myb | | Authors: | Zor, T, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the KIX Domain of CBP Bound to the Transactivation
Domain of c-Myb
J.Mol.Biol., 337, 2004
|
|
2JPR
 
 | | Joint refinement of the HIV-1 CA-NTD in complex with the assembly inhibitor CAP-1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, Gag-Pol polyprotein | | Authors: | Kelly, B.N, Kyere, S, Kinde, I, Tang, C, Howard, B.R, Robinson, H, Sundquist, W.I, Summers, M.F, Hill, C.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein
J.Mol.Biol., 373, 2007
|
|
1S4U
 
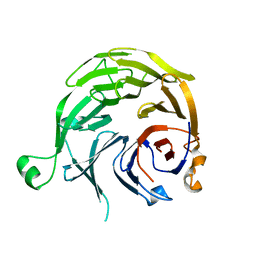 | |
2JOX
 
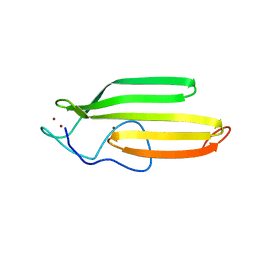 | | Embryonic Neural Inducing Factor Churchill is not a DNA-Binding Zinc Finger Protein: Solution Structure Reveals a Solvent-Exposed beta-Sheet and Zinc Binuclear Cluster | | Descriptor: | Churchill protein, ZINC ION | | Authors: | Lee, B.M, Buck-Koehntop, B.A, Martinez-Yamout, M.A, Gottesfeld, J.M, Dyson, H, Wright, P.E. | | Deposit date: | 2007-04-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed beta-Sheet and Zinc Binuclear Cluster
J.Mol.Biol., 371, 2007
|
|
2KJE
 
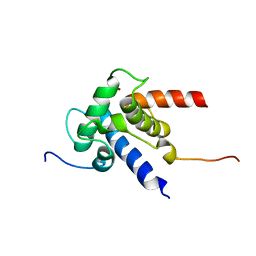 | | NMR structure of CBP TAZ2 and adenoviral E1A complex | | Descriptor: | CREB-binding protein, Early E1A 32 kDa protein, ZINC ION | | Authors: | Ferreon, J.C, Martinez-Yamout, M, Dyson, H, Wright, P.E. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1R5B
 
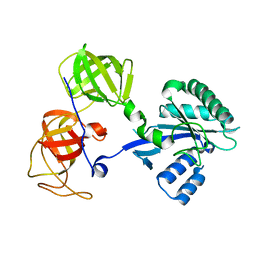 | | Crystal structure analysis of sup35 | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1R5N
 
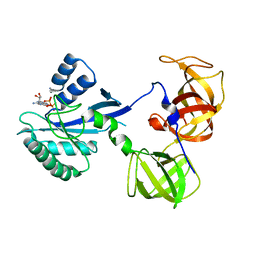 | | Crystal Structure Analysis of sup35 complexed with GDP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1FNF
 
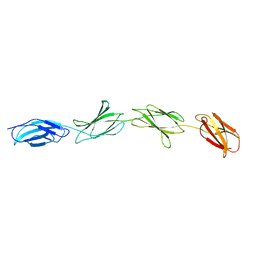 | |
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
1SV0
 
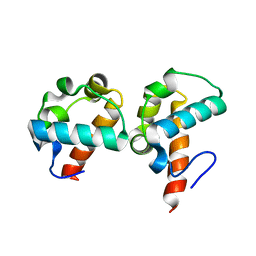 | | Crystal Structure Of Yan-SAM/Mae-SAM Complex | | Descriptor: | Ets DNA-binding protein pokkuri, modulator of the activity of Ets CG15085-PA | | Authors: | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
2L14
 
 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
1SV4
 
 | | Crystal Structure of Yan-SAM | | Descriptor: | Ets DNA-binding protein pokkuri | | Authors: | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | Deposit date: | 2004-03-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
