5YAG
 
 | | Crystal structure of mosquito arylalkylamine N-Acetyltransferase like 5b/spermine N-Acetyltransferase | | Descriptor: | AAEL004827-PA, GLYCEROL | | Authors: | Han, Q, Guan, H, Robinson, H, Li, J. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of aaNAT5b as a spermine N-acetyltransferase in the mosquito, Aedes aegypti.
PLoS ONE, 13, 2018
|
|
4D7E
 
 | | An unprecedented NADPH domain conformation in Lysine Monooxygenase NbtG from Nocardia farcinica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-LYS MONOOXYGENASE | | Authors: | Binda, C, Robinson, R, Keul, N, Rodriguez, P, Robinson, H.H, Mattevi, A, Sobrado, P. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Unprecedented Nadph Domain Conformation in Lysine Monooxygenase Nbtg Provides Insights Into Uncoupling of Oxygen Consumption from Substrate Hydroxylation.
J.Biol.Chem., 290, 2015
|
|
5YDI
 
 | | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector anopheles gambiae, new crystal packing | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Han, Q, Guan, H, Ding, H, Liao, C, Robinson, H, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
5YDH
 
 | | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector anopheles gambiae, 3.2 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Acetylcholinesterase, ... | | Authors: | Han, Q, Guan, H, Robinson, H, Ding, H, Liao, C, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
2ZJG
 
 | | Crystal structural of mouse kynurenine aminotransferase III | | Descriptor: | GLYCEROL, Kynurenine-oxoglutarate transaminase 3 | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-03-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of mouse kynurenine aminotransferase III
To be Published
|
|
4AP9
 
 | | Crystal structure of phosphoserine phosphatase from T. onnurineus in complex with NDSB-201 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Jung, T.-Y, Kim, Y.-S, Song, H.-N, Woo, E. | | Deposit date: | 2012-03-31 | | Release date: | 2012-12-26 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Identification of a Novel Ligand Binding Site in Phosphoserine Phosphatase from the Hyperthermophilic Archaeon Thermococcus Onnurineus.
Proteins, 81, 2013
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
3BRE
 
 | | Crystal Structure of P.aeruginosa PA3702 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Probable two-component response regulator | | Authors: | De, N, Pirruccello, M, Krasteva, P.V, Bae, N, Raghavan, R.V, Sondermann, H. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation-independent regulation of the diguanylate cyclase WspR.
Plos Biol., 6, 2008
|
|
6AEE
 
 | | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | | Descriptor: | 9 Mer Peptide (RL9) From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Wang, Q, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2018-08-04 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex.
Cell. Mol. Immunol., 2019
|
|
6A9B
 
 | | T4 dCMP hydroxymethylase structure solved by I-SAD using a home source | | Descriptor: | Deoxycytidylate 5-hydroxymethyltransferase, IODIDE ION, PHOSPHATE ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2018-07-12 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A cytosine modification mechanism revealed by the structure of a ternary complex of deoxycytidylate hydroxymethylase from bacteriophage T4 with its cofactor and substrate.
Iucrj, 6, 2019
|
|
3HSF
 
 | | HEAT SHOCK TRANSCRIPTION FACTOR (HSF) | | Descriptor: | HEAT SHOCK TRANSCRIPTION FACTOR | | Authors: | Damberger, F.F, Pelton, J.G, Liu, C, Cho, H, Harrison, C.J, Nelson, H.C.M, Wemmer, D.E. | | Deposit date: | 1995-08-07 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and dynamics of the DNA-binding domain of the heat shock factor from Kluyveromyces lactis.
J.Mol.Biol., 254, 1995
|
|
2VTC
 
 | | The structure of a glycoside hydrolase family 61 member, Cel61B from the Hypocrea jecorina. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CEL61B, NICKEL (II) ION | | Authors: | Karkehabadi, S, Hansson, H, Kim, S, Piens, K, Mitchinson, C, Sandgren, M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The First Structure of a Glycoside Hydrolase Family 61 Member, Cel61B from the Hypocrea Jecorina, at 1.6 A Resolution.
J.Mol.Biol., 383, 2008
|
|
2OUN
 
 | | crystal structure of PDE10A2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H, Ke, H.M. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUU
 
 | | crystal structure of PDE10A2 mutant D674A in complex with cGMP | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PW3
 
 | | Structure of the PDE4D-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The molecular basis for different recognition of substrates by phosphodiesterase families 4 and 10.
J.Mol.Biol., 371, 2007
|
|
2OUQ
 
 | | crystal structure of PDE10A2 in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUV
 
 | | crystal structure of pde10a2 mutant of D564N | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUY
 
 | | crystal structure of pde10a2 mutant D564A in complex with cAMP. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2A9A
 
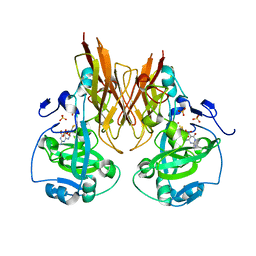 | | Crystal structure of recombinant chicken sulfite oxidase with the bound product, sulfate, at the active site | | Descriptor: | MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, SULFATE ION, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
5BZ6
 
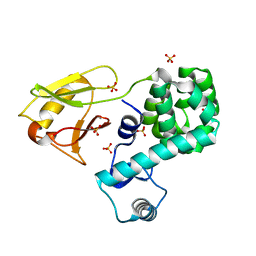 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
2OUP
 
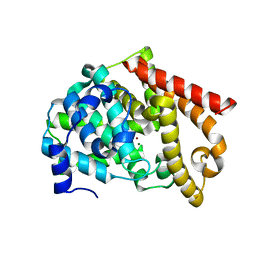 | | crystal structure of PDE10A | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2A9C
 
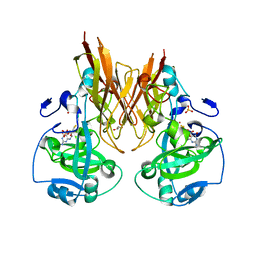 | | Crystal structure of R138Q mutant of recombinant chicken sulfite oxidase with the bound product, sulfate, at the active site | | Descriptor: | GLYCEROL, MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2A9D
 
 | | Crystal structure of recombinant chicken sulfite oxidase with Arg at residue 161 | | Descriptor: | MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, SULFATE ION, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2R62
 
 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH | | Descriptor: | Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
2R65
 
 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
