7JJM
 
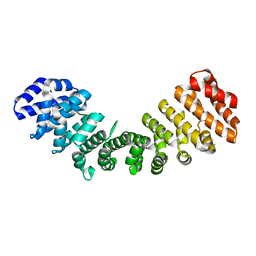 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS | | Descriptor: | CHLORIDE ION, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McGuaig, R, Tan, H.Y.A, Hardy, C, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
7RXY
 
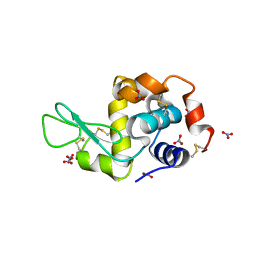 | | Hen egg-white lysozyme with ionic liquid ethylammonium nitrate 5 mol% | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Han, Q, Darmanin, C, Smith, K, Drummond, C, Greaves, T. | | Deposit date: | 2021-08-24 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing ion-binding at a protein interface: Modulation of protein properties by ionic liquids.
J Colloid Interface Sci, 650, 2023
|
|
7RYD
 
 | | Hen egg-white lysozyme with ionic liquid butylammonium nitrate 1 mol% | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Han, Q, Darmanin, C, Smith, K, Drummond, C, Greaves, T. | | Deposit date: | 2021-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Probing ion-binding at a protein interface: Modulation of protein properties by ionic liquids.
J Colloid Interface Sci, 650, 2023
|
|
7RYK
 
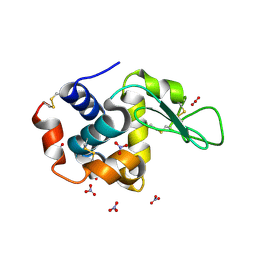 | | Hen egg-white lysozyme with ionic liquid ethanolammonium nitrate 1 mol% | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Han, Q, Darmanin, C, Smith, K, Drummond, C, Greaves, T. | | Deposit date: | 2021-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Probing ion-binding at a protein interface: Modulation of protein properties by ionic liquids.
J Colloid Interface Sci, 650, 2023
|
|
7JK7
 
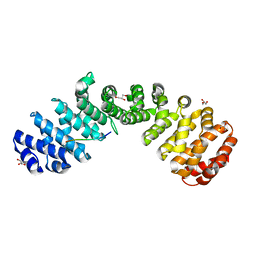 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS S111E mutant | | Descriptor: | GLYCEROL, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McCuaig, R, Tan, A.H.Y, Hardy, K, Seddiki, N, Ali, S, Dahlstron, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
2N1E
 
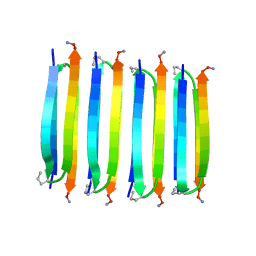 | | MAX1 peptide fibril | | Descriptor: | MAX1 peptide | | Authors: | Nagy-Smith, K, Moore, E, Schneider, J, Tycko, R. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-30 | | Method: | SOLID-STATE NMR | | Cite: | Molecular structure of monomorphic peptide fibrils within a kinetically trapped hydrogel network.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7JMU
 
 | | Hen egg-white lysozyme with ionic liquid ethylammonium nitrate | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Han, Q, Darmanin, C, Smith, K, Greaves, T, Drummond, C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Lysozyme conformational changes with ionic liquids: Spectroscopic, small angle x-ray scattering and crystallographic study.
J Colloid Interface Sci, 585, 2021
|
|
4CAU
 
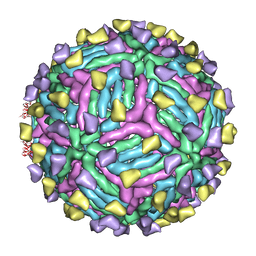 | | THREE-DIMENSIONAL STRUCTURE OF DENGUE VIRUS SEROTYPE 1 COMPLEXED WITH 2 HMAB 14C10 FAB | | Descriptor: | ENVELOPE PROTEIN E, FAB 14C10 | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A.P, Tan, T.T, Yip, A, Schul, W, Aung, M, Kostyuchenko, V.A, Leo, Y.S, Chan, S.H, Smith, K.G, Chan, A.H, Zou, G, Ooi, E.E, Kemeny, D.M, Tan, G.K, Ng, J.K, Ng, M.L, Alonso, S, Fisher, D, Shi, P.Y, Hanson, B.J, Lok, S.M, Macary, P.A. | | Deposit date: | 2013-10-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The Structural Basis for Serotype-Specific Neutralization of Dengue Virus by a Human Antibody.
Sci.Trans.Med, 4, 2012
|
|
7JJL
 
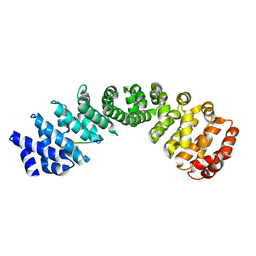 | | Crystal structure of Importin Alpha 3 in complex with human LSD1 NLS | | Descriptor: | Importin subunit alpha-3, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McCuaig, R, Tan, H.Y.A, Hardy, K, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
4BIB
 
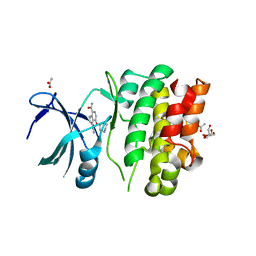 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 3-cyano-4-(piperidin-4-yloxy)-1H-indole-7-carboxamide, ACETATE ION, GLYCEROL, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4PTT
 
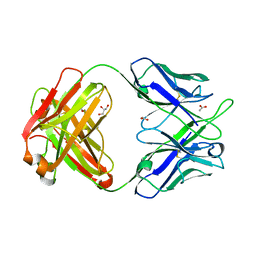 | | Crystal Structure of anti-23F strep Fab C05 | | Descriptor: | ACETATE ION, Antibody pn132p2C05, heavy chain, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Smith, K, Schrader, J.W, Pai, E.F. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4PTU
 
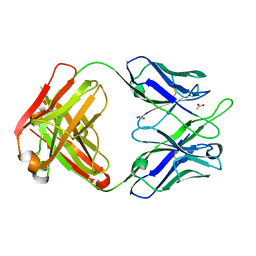 | | Crystal Structure of anti-23F strep Fab C05 with rhamnose | | Descriptor: | ACETATE ION, Antibody pn132p2C05, heavy chain, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Smith, K, Schrader, J.W, Pai, E.F. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
6BWB
 
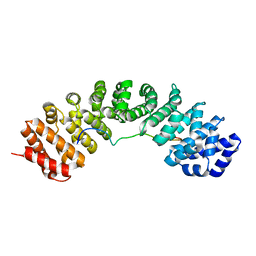 | | Hendra virus W protein C-terminus in complex with Importin alpha 3 crystal form 3 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BW1
 
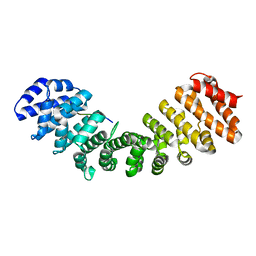 | | Hendra virus W protein C-terminus in complex with Importin alpha 1 | | Descriptor: | Importin subunit alpha-1, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
4BIE
 
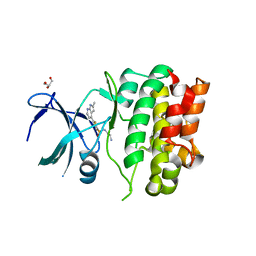 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{2-methyl-1H-pyrrolo[2,3-b]pyridin-4-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
5W41
 
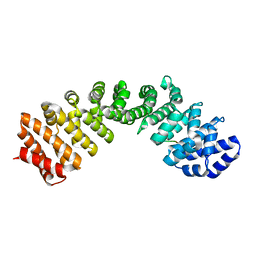 | |
1G5C
 
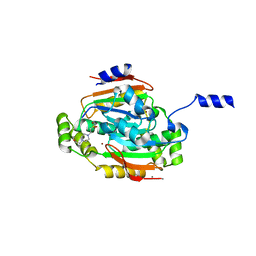 | | CRYSTAL STRUCTURE OF THE 'CAB' TYPE BETA CLASS CARBONIC ANHYDRASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-CARBONIC ANHYDRASE, CALCIUM ION, ... | | Authors: | Strop, P, Smith, K.S, Iverson, T.M, Ferry, J.G, Rees, D.C. | | Deposit date: | 2000-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the "cab"-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum.
J.Biol.Chem., 276, 2001
|
|
1E9K
 
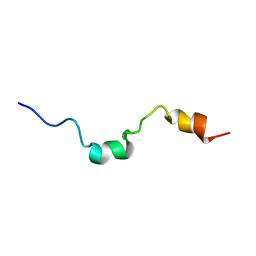 | | The structure of the RACK1 interaction sites located within the unique N-terminal region of the cAMP-specific phosphodiesterase, PDE4D5. | | Descriptor: | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Bolger, G.B, Smith, K.J, McCahill, A, Hyde, E.I, Steele, M.R, Houslay, M.D. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H NMR structural and functional characterisation of a cAMP-specific phosphodiesterase-4D5 (PDE4D5) N-terminal region peptide that disrupts PDE4D5 interaction with the signalling scaffold proteins, beta-arrestin and RACK1.
Cell. Signal., 19, 2007
|
|
4BF2
 
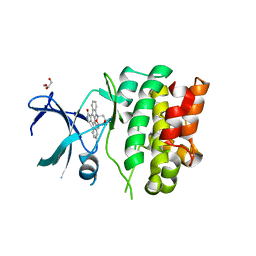 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | ACETATE ION, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-03-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BID
 
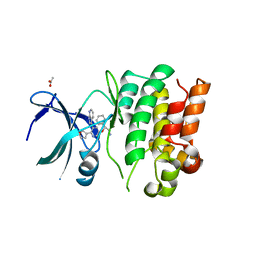 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 5-[(1R)-2-amino-1-phenylethoxy]-2-(furan-3-yl)-3-{1H-pyrrolo[2,3-b]pyridin-3-yl}pyridine, ACETATE ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BHN
 
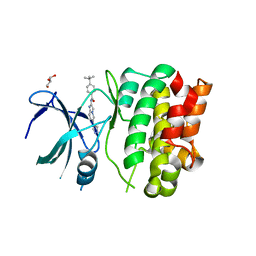 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 4-tert-butyl-N-[6-(1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIC
 
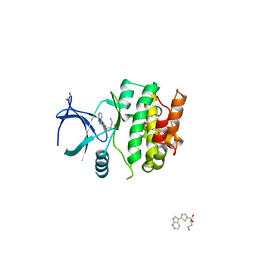 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{1H-pyrrolo[2,3-b]pyridin-3-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | Descriptor: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | Authors: | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
1AGD
 
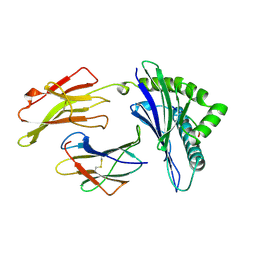 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYKL-INDEX PEPTIDE) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYKL - INDEX PEPTIDE) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGE
 
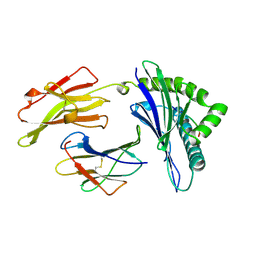 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYRL-7R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYRL - 7R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
