4U9R
 
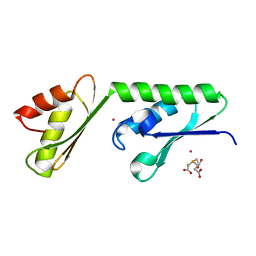 | | Structure of the N-terminal Extension from Cupriavidus metallidurans CzcP | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, CADMIUM ION, CzcP cation efflux P1-ATPase | | Authors: | Smith, A.T, Rosenzweig, A.C. | | Deposit date: | 2014-08-06 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A new metal binding domain involved in cadmium, cobalt and zinc transport.
Nat.Chem.Biol., 11, 2015
|
|
6WI6
 
 | | Crystal structure of plantacyclin B21AG | | Descriptor: | MALONATE ION, Plantacyclin B21AG | | Authors: | Smith, A.T, Gor, M.C, Vezina, B, McMahon, R, King, G, Panjikar, S, Rehm, B, Martin, J. | | Deposit date: | 2020-04-08 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and site-directed mutagenesis of circular bacteriocin plantacyclin B21AG reveals cationic and aromatic residues important for antimicrobial activity.
Sci Rep, 10, 2020
|
|
6PTE
 
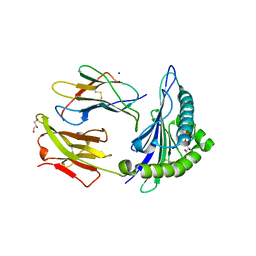 | | Crystal Structure of ILNAMITKI peptide bound to HLA-A2 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HAUS augmin-like complex subunit 3, ... | | Authors: | Smith, A.R, Arbuiso, A, Keller, G.L.J, Baker, B.M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure Based Prediction of Neoantigen Immunogenicity.
Front Immunol, 10, 2019
|
|
6VMC
 
 | |
6VM9
 
 | |
6VM8
 
 | |
6NZI
 
 | |
4ZAJ
 
 | |
6VM7
 
 | |
6VMA
 
 | |
1NPV
 
 | | Crystal structure of HIV-1 protease complexed with LDC271 | | Descriptor: | POL polyprotein, {1-BENZYL-3-[2-BENZYL-3-OXO-4-(1-OXO-1,2,3,4-TETRAHYDRO- ISOQUINOLIN-4-YL)-2,3-DIHYDRO-1H-PYRROL-2-YL]-2- HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Smith, A.B. | | Deposit date: | 2003-01-20 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors.
J.Med.Chem., 46, 2003
|
|
1U88
 
 | |
1U87
 
 | |
3GYR
 
 | | Structure of Phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, GLYCEROL, ... | | Authors: | Smith, A.W, Camara-Artigas, A, Wang, M, Francisco, W.A, Allen, J.P. | | Deposit date: | 2009-04-05 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Phenoxazinone Synthase from Streptomyces Antibioticus Reveals a New Type 2 Copper Center.
Biochemistry, 45, 2006
|
|
5AG4
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2S)-3-(3-chlorophenyl)-2-(pyridin-2-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
5AGE
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A BENZOMORPHOLINONE LIGAND | | Descriptor: | 4-[(5-methyl-1,2-oxazol-3-yl)methyl]-7-[4-(1-methylpiperidin-4-yl)butyl]-2H-1,4-benzoxazin-3(4H)-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
5AG7
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A BENZOMORPHOLINE LIGAND | | Descriptor: | GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA, ethyl (3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)acetate | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
1E61
 
 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
4GMH
 
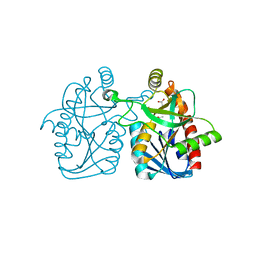 | | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ACETATE ION | | Authors: | Brown, R.L, Anderson, B.F, Norris, G.E, Tyler, P.C, Evans, G.B, Sutherland-Smith, A.J. | | Deposit date: | 2012-08-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase
To be Published
|
|
6E55
 
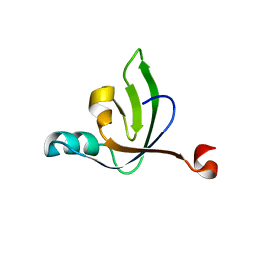 | |
8W33
 
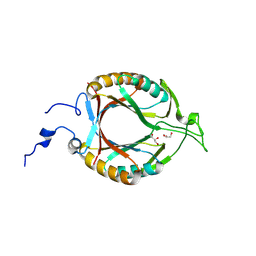 | | Structure of McrD (methyl-coenzyme M reductase operon protein D) from Methanomassiliicoccus luminyensis | | Descriptor: | GLYCEROL, McrD (methyl-coenzyme M reductase operon protein D) | | Authors: | Sutherland-Smith, A.J, Carbone, V, Schofield, L.R, Ronimus, R.S. | | Deposit date: | 2024-02-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of methanogen McrD, a methyl-coenzyme M reductase-associated protein.
Febs Open Bio, 2024
|
|
7TZI
 
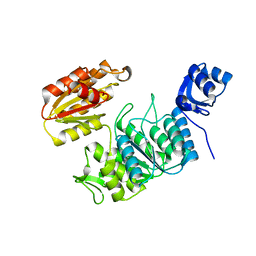 | | Structure of a pseudomurein peptide ligase type E from Methanothermobacter thermautotrophicus | | Descriptor: | Mur ligase family protein | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
7UFP
 
 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | Mur ligase middle domain protein, SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2022-03-23 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
6GNV
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a isopropyl methyl indole aryl sulphonamide ligand | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GNH
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[(4-methoxyphenyl)sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
