6F3L
 
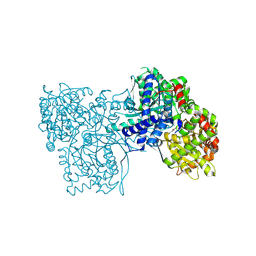 | | The crystal structure of Glycogen Phosphorylase in complex with 10b | | Descriptor: | 6-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-1~{H}-1,2,4-triazol-3-yl]naphthalene-2-carboxylic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Barkas, T.A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6F3J
 
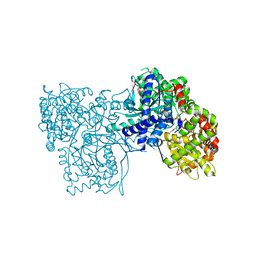 | | The crystal structure of Glycogen Phosphorylase in complex with 10a | | Descriptor: | 4-[4-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-4~{H}-1,2,4-triazol-3-yl]phenyl]benzoic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6F3U
 
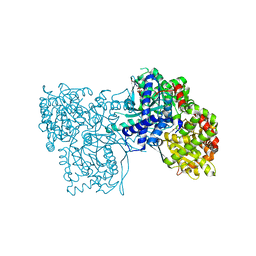 | | The crystal structure of Glycogen Phosphorylase in complex with 10h | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(5-naphthalen-1-yl-4~{H}-1,2,4-triazol-3-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Gkerdi, A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6F3R
 
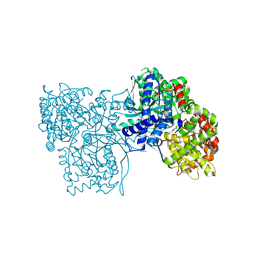 | | The crystal structure of Glycogen Phosphorylase in complex with 10c | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{R})-2-[5-(9~{H}-fluoren-2-yl)-4~{H}-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Barkas, T.A, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
6F3S
 
 | | The crystal structure of Glycogen Phosphorylase in complex with 10d | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[5-(4-phenylphenyl)-4~{H}-1,2,4-triazol-3-yl]oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
3T3D
 
 | | Glycogen phosphorylase b in complex with GlcU | | Descriptor: | 1-beta-D-glucopyranosylpyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3I
 
 | | Glycogen Phosphorylase b in complex with GlcCF3U | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3H
 
 | | Glycogen Phosphorylase b in complex with GlcIU | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-iodopyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3G
 
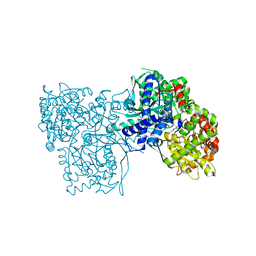 | | Glycogen Phosphorylase b in complex with GlcBrU | | Descriptor: | 5-bromo-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3E
 
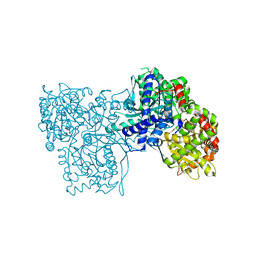 | | Glycogen phosphorylase b in complex with GlcClU | | Descriptor: | 5-chloro-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
1H5U
 
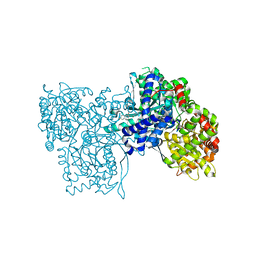 | | THE 1.76 A RESOLUTION CRYSTAL STRUCTURE OF GLYCOGEN PHOSPHORYLASE B COMPLEXED WITH GLUCOSE AND CP320626, A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Archontis, G. | | Deposit date: | 2001-05-25 | | Release date: | 2001-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The 1.76 A Resolution Crystal Structure of Glycogen Phosphorylase B Complexed with Glucose, and Cp320626, a Potential Antidiabetic Drug
Bioorg.Med.Chem., 10, 2002
|
|
4EL5
 
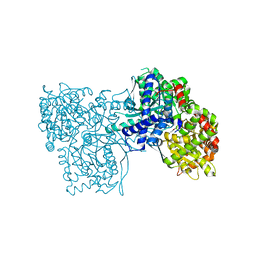 | | Crystal structure of GPb in complex with DK12 | | Descriptor: | 5-ethynyl-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EJ2
 
 | | Crystal structure of GPb in complex with DK10 | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(hept-1-yn-1-yl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
1BX3
 
 | | EFFECTS OF COMMONLY USED CRYOPROTECTANTS ON GLYCOGEN PHOSPHORYLASE ACTIVITY AND STRUCTURE | | Descriptor: | PROTEIN (GLYCOGEN PHOSPHORYLASE B), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tsitsanou, K.E, Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Gregoriou, M, Watson, K.A, Johnson, L.N, Fleet, G.W.J. | | Deposit date: | 1999-10-13 | | Release date: | 1999-10-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of commonly used cryoprotectants on glycogen phosphorylase activity and structure.
Protein Sci., 8, 1999
|
|
4EKE
 
 | | Crystal structure of GPb in complex with DK11 | | Descriptor: | 3-(beta-D-glucopyranosyl)-6-pentylfuro[2,3-d]pyrimidin-2(3H)-one, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-09 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EKY
 
 | | Crystal structure of GPb in complex with DK15 | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(pent-1-yn-1-yl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4EL0
 
 | | Crystal structure of GPb in complex with DK16 | | Descriptor: | 3-(beta-D-glucopyranosyl)-6-propylfuro[2,3-d]pyrimidin-2(3H)-one, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
1B4D
 
 | | AMIDOCARBAMATE INHIBITOR OF GLYCOGEN PHOSPHORYLASE | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE B), ... | | Authors: | Tsitsanou, K.E, Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Gregoriou, M, Watson, K.A, Johnson, L.N, Fleet, G.W.J. | | Deposit date: | 1998-12-18 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of commonly used cryoprotectants on glycogen phosphorylase activity and structure.
Protein Sci., 8, 1999
|
|
6S4H
 
 | | The crystal structure of glycogen phosphorylase in complex with 8 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-phenyl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6S52
 
 | | The crystal structure of glycogen phosphorylase in complex with 14 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(5-phenyl-1,2,3,4-tetrazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6S4O
 
 | | The crystal structure of glycogen phosphorylase in complex with 9 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-naphthalen-2-yl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6S4K
 
 | | The crystal structure of glycogen phosphorylase in complex with 12 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(4-phenyl-1,3-thiazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6S4R
 
 | | The crystal structure of glycogen phosphorylase in complex with 11 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-naphthalen-2-yl-1,3-thiazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Papaioannou, O.S.E, Solovou, T.G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6R0H
 
 | | Glycogen Phosphorylase b in complex with 3 | | Descriptor: | 3-(4-fluorophenyl)-~{N}-[(2~{R},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]benzamide, Glycogen phosphorylase, muscle form, ... | | Authors: | Tsagkarakou, S.A, Koulas, M.S, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-10 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
6R0I
 
 | | Glycogen Phosphorylase b in complex with 4 | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Koulas, M.S, Tsagkarakou, S.A, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
