4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URL
 
 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URN
 
 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | Descriptor: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
1N76
 
 | | CRYSTAL STRUCTURE OF HUMAN SEMINAL LACTOFERRIN AT 3.4 A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN, ... | | Authors: | Kumar, J, Weber, W, Munchau, S, Yadav, S, Singh, S.B, Sarvanan, K, Paramsivam, M, Sharma, S, Kaur, P, Bhushan, A, Srinivasan, A, Betzel, C, Singh, T.P. | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of human seminal lactoferrin at 3.4A resolution
Indian J.Biochem.Biophys., 40, 2003
|
|
4URO
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Novobiocin | | Descriptor: | DNA GYRASE SUBUNIT B, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
2E9E
 
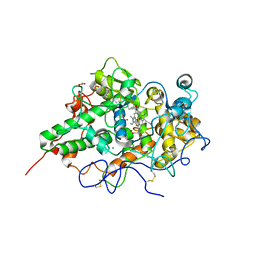 | | Crystal structure of the complex of goat lactoperoxidase with Nitrate at 3.25 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Prem kumar, R, Singh, N, Sharma, S, Singh, S.B, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with Nitrate at 3.25 A resolution
To be Published
|
|
3ERH
 
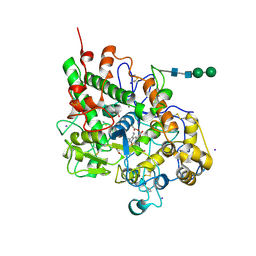 | | First structural evidence of substrate specificity in mammalian peroxidases: Crystal structures of substrate complexes with lactoperoxidases from two different species | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Sheikh, I.A, Singh, N, Singh, A.K, Sinha, M, Singh, S.B, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-10-02 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
2OJV
 
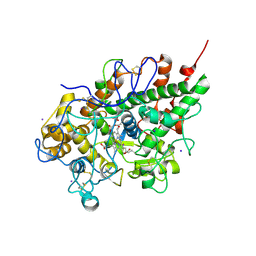 | | Crystal structure of a ternary complex of goat lactoperoxidase with cyanide and iodide ions at 2.4 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Singh, A.K, Singh, N, Singh, S.B, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-01-15 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a ternary complex of goat lactoperoxidase with cyanide and iodide ions at 2.4 A resolution
To be Published
|
|
1SOJ
 
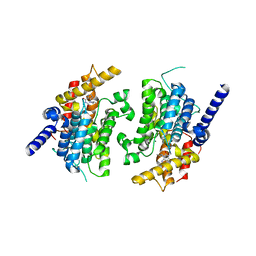 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B IN COMPLEX WITH IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, cGMP-inhibited 3',5'-cyclic phosphodiesterase B | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
1SO2
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B In COMPLEX WITH A DIHYDROPYRIDAZINE INHIBITOR | | Descriptor: | 1-DEOXY-1-[(2-HYDROXYETHYL)(NONANOYL)AMINO]HEXITOL, 6-(4-{[2-(3-IODOBENZYL)-3-OXOCYCLOHEX-1-EN-1-YL]AMINO}PHENYL)-5-METHYL-4,5-DIHYDROPYRIDAZIN-3(2H)-ONE, MAGNESIUM ION, ... | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
3D5H
 
 | | Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Singh, S.B, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-05-16 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance
To be Published
|
|
4MDT
 
 | | Structure of LpxC bound to the reaction product UDP-(3-O-(R-3-hydroxymyristoyl))-glucosamine | | Descriptor: | PHOSPHATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Clayton, G.M, Klein, D.J, Rickert, K.W, Patel, S.B, Kornienko, M, Zugay-Murphy, J, Reid, J.C, Tummala, S, Sharma, S, Singh, S.B, Miesel, L, Lumb, K.J, Soisson, S.M. | | Deposit date: | 2013-08-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of the Bacterial Deacetylase LpxC Bound to the Nucleotide Reaction Product Reveals Mechanisms of Oxyanion Stabilization and Proton Transfer.
J.Biol.Chem., 288, 2013
|
|
2I0H
 
 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1IGW
 
 | | Crystal Structure of the Isocitrate Lyase from the A219C mutant of Escherichia coli | | Descriptor: | Isocitrate lyase, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Britton, K.L, Abeysinghe, I.S.B, Baker, P.J, Barynin, V, Diehl, P, Langridge, S.J, McFadden, B.A, Sedelnikova, S.E, Stillman, T.J, Weeradechapon, K, Rice, D.W. | | Deposit date: | 2001-04-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure and domain organization of Escherichia coli isocitrate lyase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
