6CQM
 
 | | Crystal Structure of mitochondrial single-stranded DNA binding proteins from S. cerevisiae, Rim1 (Form2) | | Descriptor: | Single-stranded DNA-binding protein RIM1, mitochondrial | | Authors: | Singh, S.P, Kukshal, V, Bona, P.D, Lytle, A.K, Edwin, A, Galletto, R. | | Deposit date: | 2018-03-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.
Nucleic Acids Res., 46, 2018
|
|
6CQO
 
 | | Crystal Structure of mitochondrial single-stranded DNA binding proteins from S. cerevisiae (SeMet Labeled), Rim1 (Form2) | | Descriptor: | Single-stranded DNA-binding protein RIM1, mitochondrial | | Authors: | Singh, S.P, Kukshal, V, Bona, P.D, Lytle, A.K, Edwin, A, Galletto, R. | | Deposit date: | 2018-03-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.
Nucleic Acids Res., 46, 2018
|
|
6CQK
 
 | | Crystal Structure of mitochondrial single-stranded DNA binding proteins from S. cerevisiae, Rim1 (Form1) | | Descriptor: | SsDNA-binding protein essential for mitochondrial genome maintenance | | Authors: | Singh, S.P, Kukshal, V, Bona, P.D, Lytle, A.K, Edwin, A, Galletto, R. | | Deposit date: | 2018-03-15 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.
Nucleic Acids Res., 46, 2018
|
|
3GXO
 
 | | Structure of the Mitomycin 7-O-methyltransferase MmcR with bound Mitomycin A | | Descriptor: | CALCIUM ION, MmcR, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
5YDD
 
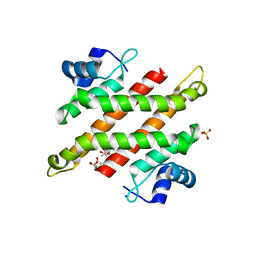 | | Crystal structure of C-terminal domain of Rv1828 from Mycobacterium tuberculosis | | Descriptor: | (6R,8S,9S)-8-(hydroxymethyl)-6,11,11-tris(oxidanyl)-9-propyl-dodecanoic acid, GLYCEROL, SODIUM ION, ... | | Authors: | Singh, S, Karthiekeyan, S. | | Deposit date: | 2017-09-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characteristics of the essential pathogenicity factor Rv1828, a MerR family transcription regulator from Mycobacterium tuberculosis.
FEBS J., 285, 2018
|
|
1KFT
 
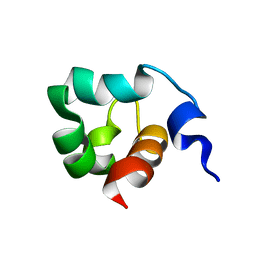 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
3GWZ
 
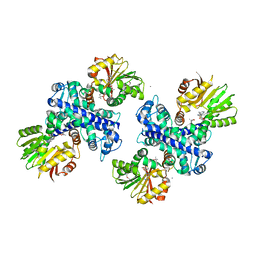 | | Structure of the Mitomycin 7-O-methyltransferase MmcR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MmcR, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
8CMU
 
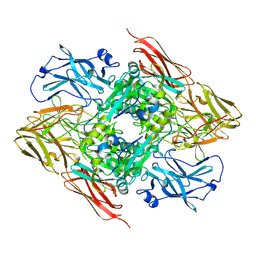 | | High resolution structure of the coagulation Factor XIII A2B2 heterotetramer complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Urgular, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | The cryo-EM structure of the plasma coagulation Factor XIII complex at 2.68 Angstrom resolution.
To Be Published
|
|
8CMT
 
 | | Structure of the plasma coagulation Factor XIII A2B2 heterotetrameric complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Ugurlar, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | The cryo-EM structure of plasma coagulation Factor XIII heterotetrameric complex at 2.68 Angstrom resolution.
To Be Published
|
|
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
7XOY
 
 | | Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine and serine. | | Descriptor: | Putative cystathionine beta-synthase Rv1077, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-05-01 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
4URL
 
 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URN
 
 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | Descriptor: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
2A5S
 
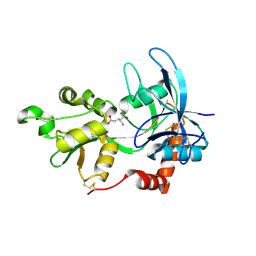 | | Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate | | Descriptor: | GLUTAMIC ACID, N-methyl-D-aspartate receptor NMDAR2A subunit | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2A5T
 
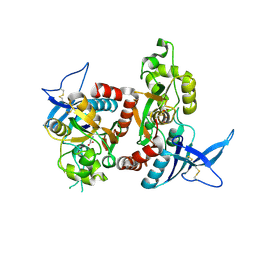 | | Crystal Structure Of The NR1/NR2A ligand-binding cores complex | | Descriptor: | GLUTAMIC ACID, GLYCINE, N-methyl-D-aspartate receptor NMDAR1-4a subunit, ... | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
7LNU
 
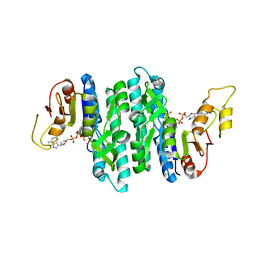 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to isopentenyl monophosphate and ATP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNW
 
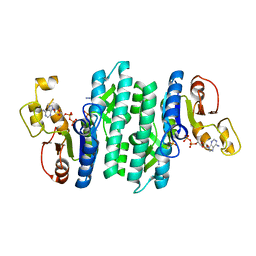 | |
7LNX
 
 | | I146A mutant of the isopentenyl phosphate kinase from Candidatus methanomethylophilus alvus | | Descriptor: | (2E)-3-methylhept-2-en-1-yl dihydrogen phosphate, (2Z)-3-methylhept-2-en-1-yl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Johnson, B.P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNT
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to benzyl monophosphate and ATP | | Descriptor: | (phenylmethyl) dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNV
 
 | | Apo Structure of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus | | Descriptor: | GLYCEROL, Isopentenyl phosphate kinase, MALONATE ION | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
3C64
 
 | | The MC179 portion of the Cysteine-rich Interdomain Region (CIDR) of a Plasmodium falciparum Erythrocyte Membrane Protein-1 (PfEMP1) | | Descriptor: | CHLORIDE ION, PfEMP1 variant 2 of strain MC, TETRAETHYLENE GLYCOL | | Authors: | Klein, M.M, Gittis, A.G, Su, H.P, Makobongo, M.O, Moore, J.M, Singh, S, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-02-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cysteine-Rich Interdomain Region from the Highly Variable Plasmodium falciparum Erythrocyte Membrane Protein-1 Exhibits a Conserved Structure.
Plos Pathog., 4, 2008
|
|
7SHE
 
 | | Cryo-EM structure of human GPR158 | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
7SHF
 
 | | Cryo-EM structure of GPR158 coupled to the RGS7-Gbeta5 complex | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
3BUJ
 
 | | Crystal Structure of CalO2 | | Descriptor: | CalO2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McCoy, J.G, Johnson, H.D, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
Proteins, 74, 2009
|
|
2OJV
 
 | | Crystal structure of a ternary complex of goat lactoperoxidase with cyanide and iodide ions at 2.4 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Singh, A.K, Singh, N, Singh, S.B, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-01-15 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a ternary complex of goat lactoperoxidase with cyanide and iodide ions at 2.4 A resolution
To be Published
|
|
