6D6Z
 
 | |
6DG3
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with caesium | | Descriptor: | CESIUM ION, PHOSPHATE ION, Pyridinium-3,5-biscarboxylic acid mononucleotide sulfurtransferase | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2018-05-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | Unique cesium-binding sites in proteins, a case study with the sacrificial sulfur transferase LarE
J Life Sci, 2021
|
|
4MSF
 
 | | Crystal structure of the complex of goat lactoperoxidase with 3-hydroxymethyl phenol at 1.98 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(hydroxymethyl)phenol, ... | | Authors: | Singh, A, Singh, R.P, Sinha, M, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with 3-hydroxymethyl phenol at 1.98 Angstrom resolution
To be published
|
|
3UBA
 
 | | Crystal structure of the complex of bovine lactoperoxidase with p-hydroxycinnamic acid at 2.6 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, A.K, Singh, R.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with p-hydroxycinnamic acid at 2.6 A resolution
to be published
|
|
4OEK
 
 | | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYLETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kumar, M, Singh, R.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A
To be Published
|
|
6B2O
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, C176A variant | | Descriptor: | ATP-utilizing enzyme of the PP-loopsuperfamily, PHOSPHATE ION, SULFATE ION | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Analysis of the Active Site Cysteine Residue of the Sacrificial Sulfur Insertase LarE from Lactobacillus plantarum.
Biochemistry, 57, 2018
|
|
6C1W
 
 | | A tethered niacin-derived pincer complex with a nickel-carbon or sulfite-carbon bond in lactate racemase | | Descriptor: | (4S)-5-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-4-sulfo-1,4-dihydropyridine-3-carbothioic S-acid, 3-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium, Lactate racemase, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2018-01-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Lactate Racemase Nickel-Pincer Cofactor Operates by a Proton-Coupled Hydride Transfer Mechanism.
Biochemistry, 57, 2018
|
|
3TGY
 
 | | Crystal structure of the complex of Bovine Lactoperoxidase with Ascorbic acid at 2.35 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Singh, R.P, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution.
Biochim.Biophys.Acta, 1865, 2017
|
|
6B2M
 
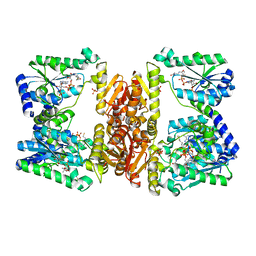 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with coenzyme A | | Descriptor: | ATP-utilizing enzyme of the PP-loopsuperfamily, COENZYME A, PHOSPHATE ION | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Analysis of the Active Site Cysteine Residue of the Sacrificial Sulfur Insertase LarE from Lactobacillus plantarum.
Biochemistry, 57, 2018
|
|
6BWR
 
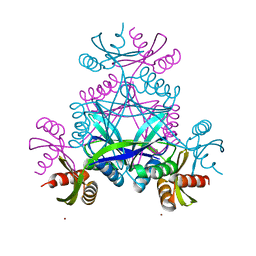 | | LarC2, the C-terminal domain of a cyclometallase involved in the synthesis of the NPN cofactor of lactate racemase, in complex with nickel | | Descriptor: | NICKEL (II) ION, Pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickel insertion protein | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Biosynthesis of the nickel-pincer nucleotide cofactor of lactate racemase requires a CTP-dependent cyclometallase.
J. Biol. Chem., 293, 2018
|
|
6BWQ
 
 | | LarC2, the C-terminal domain of a cyclometallase involved in the synthesis of the NPN cofactor of lactate racemase, in complex with MnCTP | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biosynthesis of the nickel-pincer nucleotide cofactor of lactate racemase requires a CTP-dependent cyclometallase.
J. Biol. Chem., 293, 2018
|
|
6BWO
 
 | | LarC2, the C-terminal domain of a cyclometallase involved in the synthesis of the NPN cofactor of lactate racemase, apo form | | Descriptor: | Pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickel insertion protein | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biosynthesis of the nickel-pincer nucleotide cofactor of lactate racemase requires a CTP-dependent cyclometallase.
J. Biol. Chem., 293, 2018
|
|
3SXV
 
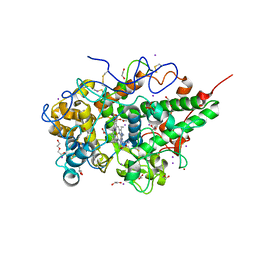 | | Crystal structure of the complex of goat lactoperoxidase with amitrole at 2.1 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with amitrole at 2.1 A resolution
TO BE PUBLISHED
|
|
6CF3
 
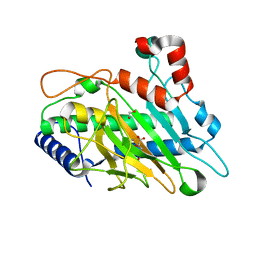 | | Ethylene forming enzyme Y306A variant in complex with manganese and 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 2024
|
|
3NAK
 
 | | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite at 3.3 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vikram, G, Singh, R.P, Singh, A.K, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite at 3.3 A resolution
To be Published
|
|
3QF1
 
 | | Crystal structure of the complex of caprine lactoperoxidase with diethylenediamine at 2.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-01-21 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of caprine lactoperoxidase with diethylenediamine at 2.6A resolution
To be Published
|
|
3RKE
 
 | | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution | | Descriptor: | (4-aminophenyl)-imidazol-1-yl-methanone, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dube, D, Singh, R.P, Sinha, M, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution
TO BE PUBLISHED
|
|
3R5O
 
 | | Crystal structure of the complex of bovine lactoperoxidase with 4-allyl-2-methoxyphenol at 2.6 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, A.K, Singh, R.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-19 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with 4-allyl-2-methoxyphenol at 2.6 A resolution
To be Published
|
|
3R4X
 
 | | Crystal structure of bovine lactoperoxidase complexed with pyrazine-2-carboxamide at 2 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of bovine lactoperoxidase complexed with Pyrazine-2-carboxamide at 2 A resolution
To be Published
|
|
2KAU
 
 | | THE CRYSTAL STRUCTURE OF UREASE FROM KLEBSIELLA AEROGENES AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | NICKEL (II) ION, UREASE (ALPHA CHAIN), UREASE (BETA CHAIN), ... | | Authors: | Jabri, E, Carr, M.B, Hausinger, R.P, Karplus, P.A. | | Deposit date: | 1995-02-16 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of urease from Klebsiella aerogenes.
Science, 268, 1995
|
|
1A5O
 
 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5N
 
 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5K
 
 | | K217E VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5L
 
 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5M
 
 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
