7PD5
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 in complex with 4-aminobenzoic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 in complex with 4-aminobenzoic acid
To Be Published
|
|
7QIB
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP
To Be Published
|
|
7QOQ
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UMP and Magnesium | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | Authors: | Silva, A, Barbosa Pereira, P.J, Macedo-Ribeiro, S, Costa, D. | | Deposit date: | 2021-12-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UMP and Magnesium
To Be Published
|
|
5M2E
 
 | |
7PE4
 
 | |
7P8G
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 - apo form | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MALONATE ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-07-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 - apo form
To Be Published
|
|
7PDO
 
 | |
7PVL
 
 | |
7QCP
 
 | |
7QI9
 
 | |
7P5L
 
 | |
8P98
 
 | | BtuB3G3 bound to cyanocobalamin with ordered EL8 | | Descriptor: | CYANOCOBALAMIN, Putative surface layer protein, Vitamin B12 transporter BtuB | | Authors: | Silale, A, Abellon-Ruiz, J, van den Berg, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
8BLW
 
 | |
8P97
 
 | |
7O3D
 
 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
7O6B
 
 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
8BYT
 
 | |
8BZ2
 
 | |
1NBS
 
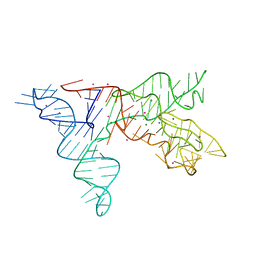 | | Crystal structure of the specificity domain of Ribonuclease P RNA | | Descriptor: | LEAD (II) ION, MAGNESIUM ION, RIBONUCLEASE P RNA | | Authors: | Krasilnikov, A.S, Yang, X, Pan, T, Mondragon, A. | | Deposit date: | 2002-12-03 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the specificity domain of Ribonuclease P
Nature, 421, 2003
|
|
8A8C
 
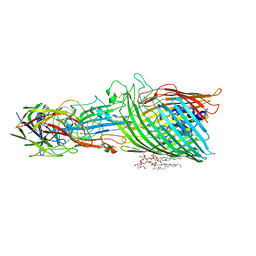 | | T5 phage receptor-binding protein pb5 bound to ferrichrome transporter FhuA | | Descriptor: | Ferrichrome outer membrane transporter/phage receptor, Receptor-binding protein pb5, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate | | Authors: | Silale, A, van den Berg, B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for host recognition and superinfection exclusion by bacteriophage T5.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1U9S
 
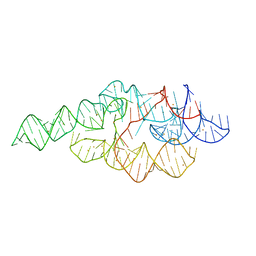 | | Crystal structure of the specificity domain of Ribonuclease P of the A-type | | Descriptor: | BARIUM ION, RIBONUCLEASE P | | Authors: | Krasilnikov, A.S, Xiao, Y, Pan, T, Mondragon, A. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Basis for structural diversity in homologous RNAs.
Science, 306, 2004
|
|
8BYM
 
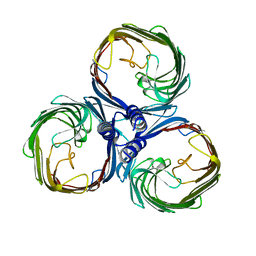 | |
8BYS
 
 | |
1LU5
 
 | | 2.4 Angstrom Crystal Structure of the Asymmetric Platinum Complex {Pt(ammine)(cyclohexylamine)}2+ Bound to a Dodecamer DNA Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', CIS-(AMMINE)(CYCLOHEXYLAMINE)PLATINUM(II) COMPLEX | | Authors: | Silverman, A.P, Bu, W, Cohen, S.M, Lippard, S.J. | | Deposit date: | 2002-05-21 | | Release date: | 2002-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4-A Crystal Structure of the Asymmetric Platinum Complex {Pt(ammine)(cyclohexylamine)}2+ Bound to
a Dodecamer DNA Duplex
J.Biol.Chem., 277, 2002
|
|
6G3U
 
 | | Structure of Pseudomonas aeruginosa Isocitrate Dehydrogenase, IDH | | Descriptor: | 2-OXOGLUTARIC ACID, Isocitrate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Crousilles, A, Welch, M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Gluconeogenic precursor availability regulates flux through the glyoxylate shunt inPseudomonas aeruginosa.
J. Biol. Chem., 293, 2018
|
|
