4PL5
 
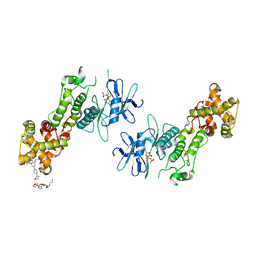 | | Crystal structure of murine IRE1 in complex with OICR573 inhibitor | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 3-methoxy-5-methyl-4'-(morpholin-4-yl)biphenyl-4-ol, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
4O1P
 
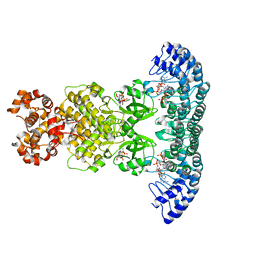 | | Crystal Structure of RNase L in complex with 2-5A and AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribonuclease L, ... | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4QLB
 
 | | Structural Basis for the Recruitment of Glycogen Synthase by Glycogenin | | Descriptor: | GLYCEROL, Probable glycogen [starch] synthase, Protein GYG-1, ... | | Authors: | Zeqiraj, E, Judd, A, Sicheri, F. | | Deposit date: | 2014-06-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recruitment of glycogen synthase by glycogenin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3TWX
 
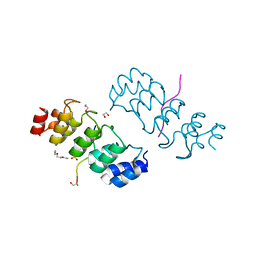 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human FNBP1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3V7D
 
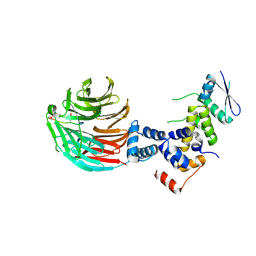 | | Crystal Structure of ScSkp1-ScCdc4-pSic1 peptide complex | | Descriptor: | Cell division control protein 4, Protein SIC1, Suppressor of kinetochore protein 1 | | Authors: | Tang, X, Orlicky, S, Mittag, T, Csizmok, V, Pawson, T, Forman-Kay, J, Sicheri, F, Tyers, M. | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TWV
 
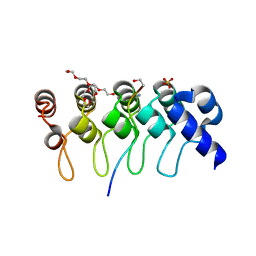 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human NUMA1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TWU
 
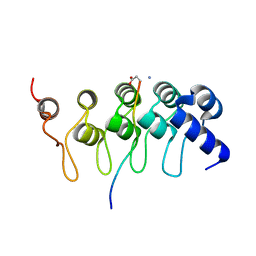 | |
6DRM
 
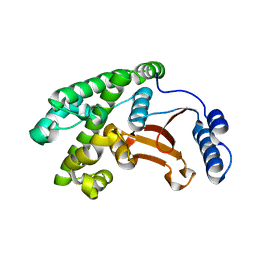 | | OTU domain of Fam105A | | Descriptor: | Inactive ubiquitin thioesterase FAM105A | | Authors: | Ceccarelli, D.F, Sicheri, F, Cordes, S. | | Deposit date: | 2018-06-12 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | FAM105A/OTULINL Is a Pseudodeubiquitinase of the OTU-Class that Localizes to the ER Membrane.
Structure, 27, 2019
|
|
3TWR
 
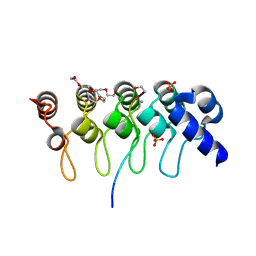 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human 3BP2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SH3 domain-binding protein 2, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TWQ
 
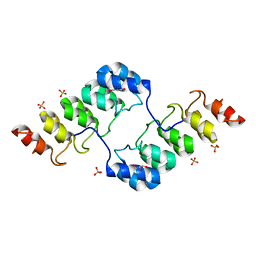 | |
3TWW
 
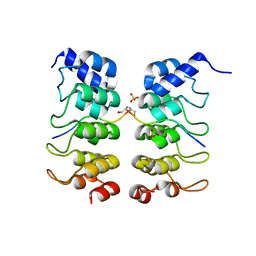 | |
3TWS
 
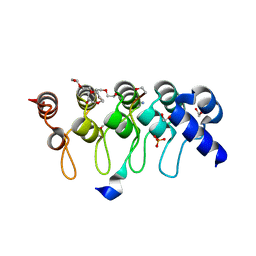 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human TERF1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TZM
 
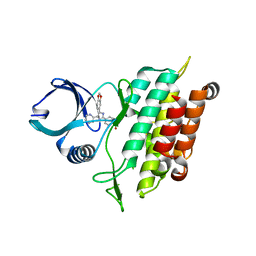 | | TGF-beta Receptor type 1 in complex with SB431542 | | Descriptor: | 4-[5-(1,3-benzodioxol-5-yl)-4-(pyridin-2-yl)-1H-imidazol-2-yl]benzamide, TGF-beta receptor type-1 | | Authors: | Ogunjimi, A.A, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Specificity of TGFbeta Family Receptor Small Molecule Inhibitors
Cell Signal, 24, 2012
|
|
3TWT
 
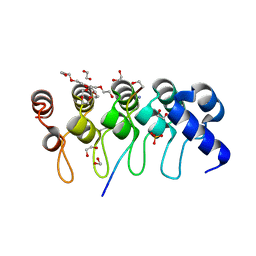 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human MCL1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, NONAETHYLENE GLYCOL, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
2QGX
 
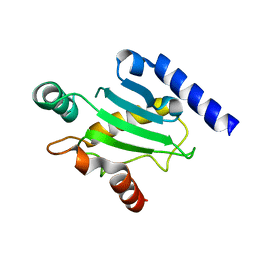 | | Ubiquitin-conjugating enzyme E2Q | | Descriptor: | Ubiquitin-conjugating enzyme E2 Q1 | | Authors: | Neculai, D, Avvakumov, G.V, Xue, S, Walker, J.R, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Sicheri, F, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
2OB4
 
 | | Human Ubiquitin-Conjugating Enzyme CDC34 | | Descriptor: | Ubiquitin-conjugating enzyme E2-32 kDa complementing | | Authors: | Neculai, D, Avvakumov, G.V, Xue, S, Walker, J.R, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Sicheri, F, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-18 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
