1TQ9
 
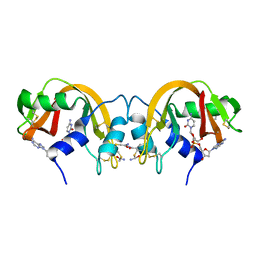 | | Non-covalent swapped dimer of Bovine Seminal Ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, Ribonuclease, seminal | | Authors: | Sica, F, Di Fiore, A, Merlino, A, Mazzarella, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Stability of the Non-covalent Swapped Dimer of Bovine Seminal Ribonuclease: AN ENZYME TAILORED TO EVADE RIBONUCLEASE PROTEIN INHIBITOR
J.Biol.Chem., 279, 2004
|
|
2EFF
 
 | |
2P1X
 
 | |
7BFL
 
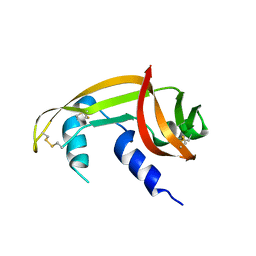 | | X-ray structure of SS-RNase-2 des116-120 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
7BFK
 
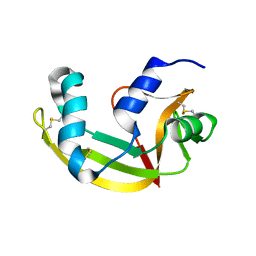 | | X-ray structure of SS-RNase-2 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
1N3Z
 
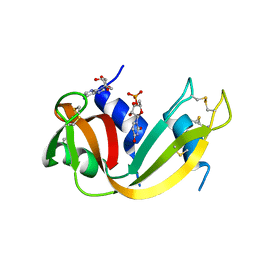 | | Crystal structure of the [S-carboxyamidomethyl-Cys31, S-carboxyamidomethyl-Cys32] monomeric derivative of the bovine seminal ribonuclease in the liganded state | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, ADENOSINE, Ribonuclease, ... | | Authors: | Sica, F, Di Fiore, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2002-10-30 | | Release date: | 2003-08-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The unswapped chain of bovine seminal ribonuclease: Crystal structure of the free and liganded monomeric derivative
Proteins, 52, 2003
|
|
1N1X
 
 | | Crystal Structure Analysis of the monomeric [S-carboxyamidomethyl-Cys31, S-carboxyamidomethyl-Cys32] Bovine seminal ribonuclease | | Descriptor: | Ribonuclease, seminal | | Authors: | Sica, F, Di Fiore, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2002-10-21 | | Release date: | 2003-08-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The unswapped chain of bovine seminal ribonuclease: Crystal structure of the free and liganded monomeric derivative
Proteins, 52, 2003
|
|
3LJF
 
 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
3BCP
 
 | | Crystal Structure of The Swapped non covalent form of P19A/L28Q/N67D BS-RNase | | Descriptor: | Seminal ribonuclease | | Authors: | Merlino, A, Ercole, C, Picone, D, Pizzo, E, Mazzarella, L, Sica, F. | | Deposit date: | 2007-11-13 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The buried diversity of bovine seminal ribonuclease: shape and cytotoxicity of the swapped non-covalent form of the enzyme
J.Mol.Biol., 376, 2008
|
|
3BCM
 
 | | Crystal Structure of The Unswapped Form of P19A/L28Q/N67D BS-RNase | | Descriptor: | PHOSPHATE ION, Seminal ribonuclease | | Authors: | Merlino, A, Ercole, C, Picone, D, Pizzo, E, Mazzarella, L, Sica, F. | | Deposit date: | 2007-11-13 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The buried diversity of bovine seminal ribonuclease: shape and cytotoxicity of the swapped non-covalent form of the enzyme
J.Mol.Biol., 376, 2008
|
|
3LJD
 
 | | The X-ray structure of zebrafish RNase1 from a new crystal form at pH 4.5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase1 | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
3BCO
 
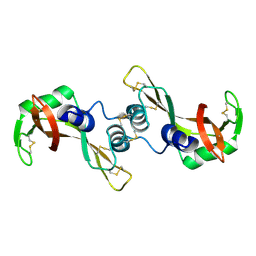 | | Crystal Structure of The Swapped FOrm of P19A/L28Q/N67D BS-RNase | | Descriptor: | Seminal ribonuclease | | Authors: | Merlino, A, Ercole, C, Picone, D, Pizzo, E, Mazzarella, L, Sica, F. | | Deposit date: | 2007-11-13 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The buried diversity of bovine seminal ribonuclease: shape and cytotoxicity of the swapped non-covalent form of the enzyme
J.Mol.Biol., 376, 2008
|
|
5EW1
 
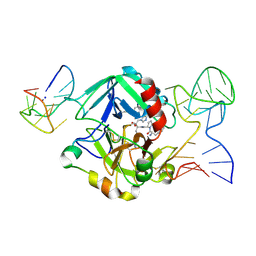 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT3, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
5EW2
 
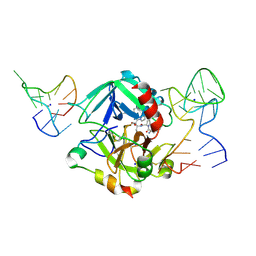 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT12, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
7NTU
 
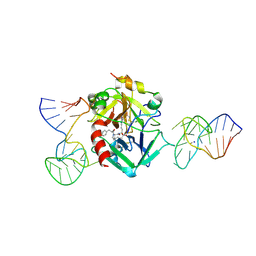 | | X-ray structure of the complex between human alpha thrombin and two duplex/quadruplex aptamers: NU172 and HD22_27mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD22_27mer, ... | | Authors: | Troisi, R, Santamaria, A, Sica, F. | | Deposit date: | 2021-03-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and functional analysis of the simultaneous binding of two duplex/quadruplex aptamers to human alpha-thrombin.
Int.J.Biol.Macromol., 181, 2021
|
|
4I7Y
 
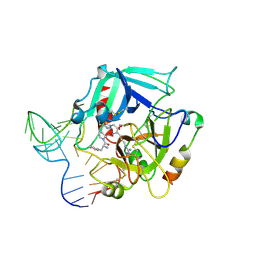 | | Crystal Structure of Human Alpha Thrombin in Complex with a 27-mer Aptamer Bound to Exosite II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, DNA (27-MER), ... | | Authors: | Pica, A, Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-12-01 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1Y92
 
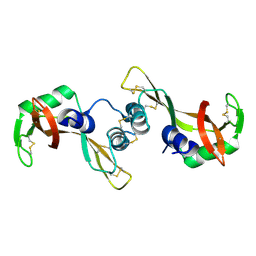 | | Crystal structure of the P19A/N67D Variant Of Bovine seminal Ribonuclease | | Descriptor: | Seminal ribonuclease | | Authors: | Picone, D, Di Fiore, A, Ercole, C, Franzese, M, Sica, F, Tomaselli, S, Mazzarella, L. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of the Hinge Loop in Domain Swapping: THE SPECIAL CASE OF BOVINE SEMINAL RIBONUCLEASE.
J.Biol.Chem., 280, 2005
|
|
1Y94
 
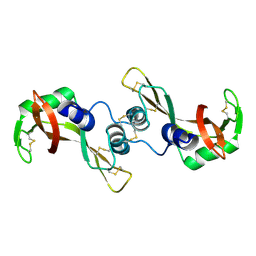 | | Crystal structure of the G16S/N17T/P19A/S20A/N67D Variant Of Bovine seminal Ribonuclease | | Descriptor: | Seminal ribonuclease | | Authors: | Picone, D, Di Fiore, A, Ercole, C, Franzese, M, Sica, F, Tomaselli, S, Mazzarella, L. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of the Hinge Loop in Domain Swapping: THE SPECIAL CASE OF BOVINE SEMINAL RIBONUCLEASE.
J.Biol.Chem., 280, 2005
|
|
6GN7
 
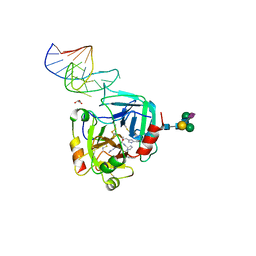 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of sodium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
11BG
 
 | | A POTENTIAL ALLOSTERIC SUBSITE GENERATED BY DOMAIN SWAPPING IN BOVINE SEMINAL RIBONUCLEASE | | Descriptor: | PROTEIN (BOVINE SEMINAL RIBONUCLEASE), SULFATE ION, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Adinolfi, S, Sica, F, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 1999-03-11 | | Release date: | 1999-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potential allosteric subsite generated by domain swapping in bovine seminal ribonuclease.
J.Mol.Biol., 293, 1999
|
|
7ZKN
 
 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
5CMX
 
 | | X-ray structure of the complex between human alpha thrombin and a duplex/quadruplex 31-mer DNA aptamer | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, RE31, ... | | Authors: | Russo Krauss, I, Pica, A, Napolitano, V, Sica, F. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding.
Nucleic Acids Res., 44, 2016
|
|
6EVV
 
 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of potassium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2017-11-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
7ZKO
 
 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form delta | | Descriptor: | 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
7ZKM
 
 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
