6M8Z
 
 | | Crystal structure of human DJ-1 without a modification on Cys-106 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Shumilin, I.A, Shabalin, I.G, Shumilina, S.V, Werenskjold, C, Utepbergenov, D, Minor, W. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A transient post-translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ-1.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6E5Z
 
 | |
6HCD
 
 | | Structure of universal stress protein from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | Authors: | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-08-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
1QR7
 
 | | CRYSTAL STRUCTURE OF PHENYLALANINE-REGULATED 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE FROM ESCHERICHIA COLI COMPLEXED WITH PB2+ AND PEP | | Descriptor: | LEAD (II) ION, PHENYLALANINE-REGULATED 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE, PHOSPHOENOLPYRUVATE, ... | | Authors: | Shumilin, I.A, Kretsinger, R.H, Bauerle, R.H. | | Deposit date: | 1999-06-18 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
1VS1
 
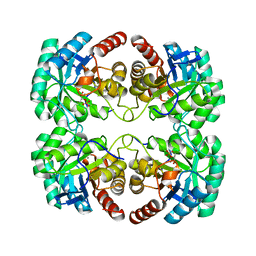 | | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Aeropyrum pernix in complex with Mn2+ and PEP | | Descriptor: | 3-deoxy-7-phosphoheptulonate synthase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE | | Authors: | Shumilin, I.A, Zhou, L, Wu, J, Woodard, R.W, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2006-03-09 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Aeropyrum pernix in complex with Mn2+ and PEP
TO BE PUBLISHED
|
|
1N8F
 
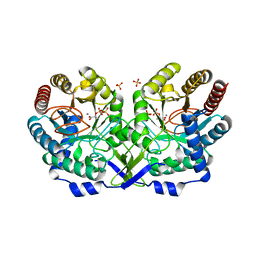 | | Crystal structure of E24Q mutant of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Escherichia Coli in complex with Mn2+ and PEP | | Descriptor: | DAHP Synthetase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Shumilin, I.A, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2002-11-20 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The High-Resolution Structure of 3-Deoxy-D-arabino-heptulosonate-7-phosphate
Synthase Reveals a Twist in the Plane of Bound Phosphoenolpyruvate
Biochemistry, 42, 2003
|
|
2DG2
 
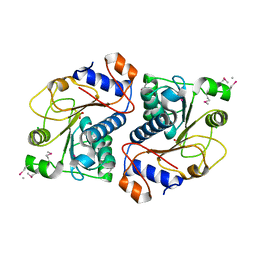 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein | | Descriptor: | Apolipoprotein A-I binding protein, CHLORIDE ION, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Zheng, H, Chruszcz, M, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and structural characterization of apolipoprotein A-I binding protein, a novel phosphoprotein with a potential role in sperm capacitation.
Endocrinology, 149, 2008
|
|
1RZM
 
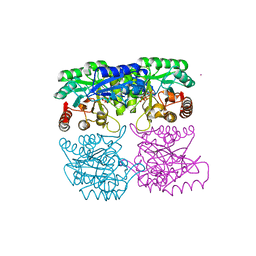 | | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHPS) from Thermotoga maritima complexed with Cd2+, PEP and E4P | | Descriptor: | CADMIUM ION, ERYTHOSE-4-PHOSPHATE, PHOSPHOENOLPYRUVATE, ... | | Authors: | Shumilin, I.A, Bauerle, R, Wu, J, Woodard, R.W, Kretsinger, R.H. | | Deposit date: | 2003-12-24 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Reaction Complex of 3-Deoxy-d-arabino-heptulosonate-7-phosphate Synthase from Thermotoga maritima Refines the Catalytic Mechanism and Indicates a New Mechanism of Allosteric Regulation.
J.Mol.Biol., 341, 2004
|
|
2O8N
 
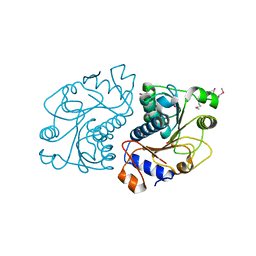 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein | | Descriptor: | ApoA-I binding protein, CHLORIDE ION, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Zheng, H, Chruszcz, M, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2006-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Characterization of Apolipoprotein A-I Binding Protein, a Novel Phosphoprotein with a Potential Role in Sperm Capacitation.
Endocrinology, 149, 2008
|
|
1KFL
 
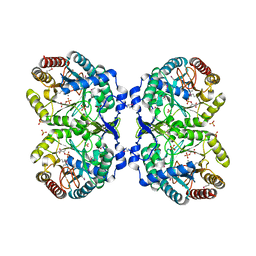 | | Crystal structure of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from E.coli complexed with Mn2+, PEP, and Phe | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Shumilin, I.A, Zhao, C, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2001-11-21 | | Release date: | 2002-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric inhibition of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase alters the coordination of both substrates.
J.Mol.Biol., 320, 2002
|
|
2OFY
 
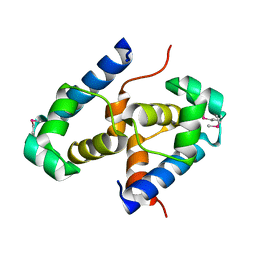 | | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | Putative XRE-family transcriptional regulator | | Authors: | Shumilin, I.A, Skarina, T, Onopriyenko, O, Yim, V, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
3H5Q
 
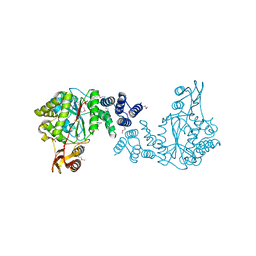 | | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus | | Descriptor: | Pyrimidine-nucleoside phosphorylase, SULFATE ION, THYMIDINE | | Authors: | Shumilin, I.A, Zimmerman, M, Cymborowski, M, Skarina, T, Onopriyenko, O, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-26 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3H3M
 
 | | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica | | Descriptor: | Flagellar protein FliT, UNKNOWN | | Authors: | Shumilin, I.A, Wang, S, Chruszcz, M, Xu, X, Le, B, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-16 | | Release date: | 2009-04-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica
To be Published
|
|
4WY2
 
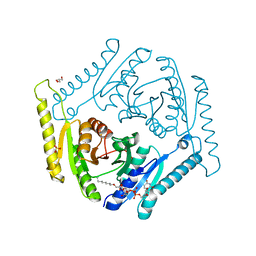 | | Crystal structure of universal stress protein E from Proteus mirabilis in complex with UDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Shumilin, I.A, Shabalin, I.G, Handing, K.B, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of universal stress protein E from Proteus mirabilis incomplex withUDP-3-O-[(3R)-3-hydroxytetradecanoyl]-N-acetyl-alpha-glucosamine
to be published
|
|
4X9P
 
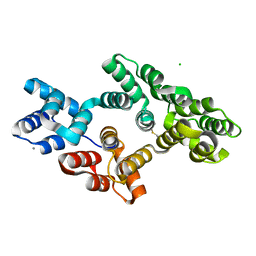 | | Crystal structure of bovine Annexin A2 | | Descriptor: | Annexin A2, CALCIUM ION, CHLORIDE ION | | Authors: | Shumilin, I.A, Hollas, H, Vedeler, A, Kretsinger, R.H. | | Deposit date: | 2014-12-11 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The native structure of annexin A2 peptides in hydrophilic environment determines their anti-angiogenic effects.
Biochem. Pharmacol., 95, 2015
|
|
4JRR
 
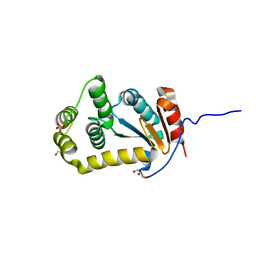 | | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Shumilin, I.A, Jameson-Lee, M, Cymborowski, M, Domagalski, M.J, Chertihin, O, Kpadeh, Z.Z, Yeh, A.J, Hoffman, P.S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila
To be Published
|
|
4K2H
 
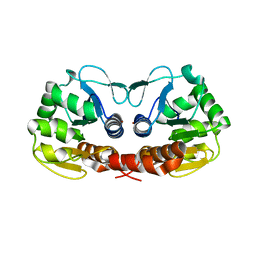 | | Crystal structure of C103A mutant of DJ-1 superfamily protein STM1931 from Salmonella typhimurium | | Descriptor: | Intracellular protease/amidase, ZINC ION | | Authors: | Shumilin, I.A, Niedzialkowska, E, Domagalski, M.J, Stam, J, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of C103A mutant of DJ-1 superfamily protein STM1931 from Salmonella typhimurium
TO BE PUBLISHED
|
|
3RO7
 
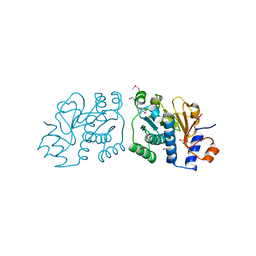 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymine. | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMINE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-25 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSF
 
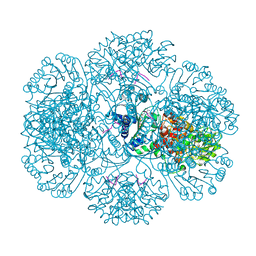 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P4-Di(adenosine-5') tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RPZ
 
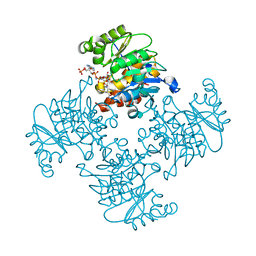 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with NADPH | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTB
 
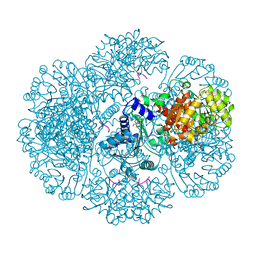 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Adenosine-3'-5'-Diphosphate | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRF
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTD
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH and ADP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSS
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTC
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NAD and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
