1V8Q
 
 | | Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TT0826 | | Authors: | Wang, H, Takemoto-Hori, C, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-13 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
1WN9
 
 | | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8 | | Descriptor: | ACETIC ACID, the hypothetical protein (TT1805) | | Authors: | Pioszak, A.A, Kishishita, S, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8
To be Published
|
|
1WXX
 
 | | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein TTHA1280 | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-02 | | Release date: | 2005-08-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1WDE
 
 | | Crystal structure of the conserved hypothetical protein APE0931 from Aeropyrum pernix K1 | | Descriptor: | Probable diphthine synthase | | Authors: | Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two archaeal diphthine synthases: insights into the post-translational modification of elongation factor 2.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1WNA
 
 | | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8 | | Descriptor: | the hypothetical protein (TT1805) | | Authors: | Pioszak, A.A, Kishishita, S, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8
To be Published
|
|
1WWS
 
 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | CALCIUM ION, hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
to be published
|
|
1WXW
 
 | | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | | Descriptor: | HEXANE-1,6-DIOL, hypothetical protein TTHA1280 | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1WWI
 
 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
1WD7
 
 | | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-2 crystal) | | Descriptor: | Uroporphyrinogen III Synthase | | Authors: | Mizohata, E, Matsuura, T, Sakai, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-12 | | Release date: | 2004-11-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-2 crystal)
to be published
|
|
1WDZ
 
 | | Crystal structure of RCB domain of IRSp53 | | Descriptor: | insulin receptor substrate p53 | | Authors: | Murayama, K, Suetsugu, S, Seto, A, Shirouzu, M, Takenawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-21 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of RCB domain of IRSp53
TO BE PUBLISHED
|
|
1WG8
 
 | |
1WCX
 
 | | Crystal Structure of Mutant Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (L75M/I193M/L248M, SeMet derivative, Form-1 crystal) | | Descriptor: | GLYCEROL, Uroporphyrinogen III Synthase | | Authors: | Mizohata, E, Matsuura, T, Murayama, K, Sakai, H, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-06 | | Release date: | 2005-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from Thermus thermophilus HB8
To be Published
|
|
1WK4
 
 | | Crystal structure of ttk003001606 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ttk003001606 | | Authors: | Kaminishi, T, Sakai, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-30 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ttk003001606
To be Published
|
|
1WJX
 
 | | Crystal sturucture of TT0801 from Thermus thermophilus | | Descriptor: | POTASSIUM ION, SsrA-binding protein | | Authors: | Bessho, Y, Shibata, R, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7X73
 
 | | Structure of G9a in complex with RK-701 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nat Commun, 14, 2023
|
|
3VHL
 
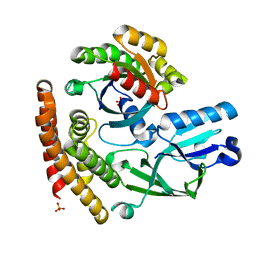 | | Crystal structure of the DHR-2 domain of DOCK8 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 8, PHOSPHATE ION | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Nishizak, T, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-08-26 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses.
Blood, 119, 2012
|
|
3VJO
 
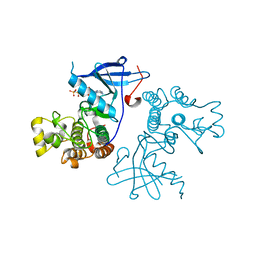 | | Crystal structure of the wild-type EGFR kinase domain in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
7DDQ
 
 | | Structure of RC-LH1-PufX from Rhodobacter veldkampii | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Antenna pigment protein alpha chain, Antenna pigment protein beta chain, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2020-10-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the photosynthetic RC-LH1-PufX supercomplex at 2.8-angstrom resolution.
Sci Adv, 7, 2021
|
|
7XN1
 
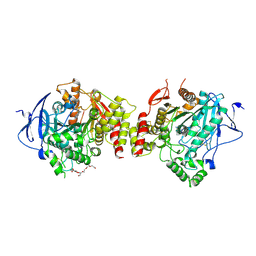 | | Crystal structure of human acetylcholinesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase, ... | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
7Y4A
 
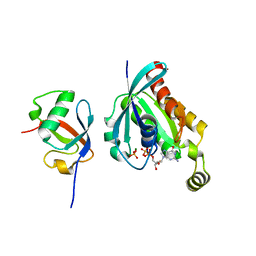 | | Crystal structure of human ELMO1 RBD-RhoG complex | | Descriptor: | Engulfment and cell motility protein 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tsuda, K, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Targeting Ras-binding domain of ELMO1 by computational nanobody design.
Commun Biol, 6, 2023
|
|
7EG2
 
 | | Crystal structure of the apoAequorin complex with (S)-daCTZ | | Descriptor: | (2~{S})-2-(hydroxymethyl)-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-3~{H}-inden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7EG3
 
 | | Crystal structure of the apoAequorin complex with (S)-HM-daCTZ | | Descriptor: | (2~{S})-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-2,3-dihydroinden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7CPJ
 
 | | ycbZ-stalled 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shirouzu, M, Ito, T. | | Deposit date: | 2020-08-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The landscape of translational stall sites in bacteria revealed by monosome and disome profiling.
Rna, 28, 2022
|
|
3VA2
 
 | | Crystal structure of human Interleukin-5 in complex with its alpha receptor | | Descriptor: | Interleukin-5, Interleukin-5 receptor subunit alpha | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-12-28 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural basis of interleukin-5 dimer recognition by its alpha receptor
Protein Sci., 21, 2012
|
|
